Single cell analysis enables researchers to gain novel insights across a diverse set of research areas, including developmental biology, tumor heterogeneity, and disease pathogenesis and progression. It permits comparisons of the transcriptomes of individual cells and allows assessment of transcriptional similarities and differences within a population of cells. Because of technological breakthroughs such as single cell isolation, whole transcriptome amplification (WTA), and NGS library preparation, experiments using single cells are now possible – opening a wealth of exciting new insights for you to discover.
Quick Biology’s single cell/low-input RNA-seq (scRNA-seq) service enables the reliable investigation of the transcriptome from a single cell to a low amount of RNA with minimal bias. It also allows you to uncover more of the transcriptome with the same sequencing depth, and delivers data including both mRNA and lncRNA, enabling the analysis of this important class of regulatory RNAs.
Request a scRNA-Seq Quote, Start Here!
Quick Biology’s single cell / low-input RNA-seq service (scRNA-seq) delivers:
· Transcript discovery and differential expression from single eukaryotic cells and RNA-seq from limited samples including viral RNA.
· An all-in-one, direct cell-to-library solution incorporating robust whole transcriptome amplification (WTA) and highly efficient library preparations
· Completely PCR-free cell-to-library protocol to minimize bias and maximize transcript detection
· Libraries from single cells using a Quick Biology optimized streamlined protocol
· RNA-seq data includes both mRNA and lincRNA
· Fewer sequence errors – this is perfect for viral genome sequencing
Our scRNA-seq is suitable for transcriptome amplification for the analysis of:
(1) mRNA with poly A+ tails
(2) Total RNA
Note: Not recommended for tRNAs, miRNAs, severely degraded RNA, RNA from FFPE material or samples fixed by formaldehyde, glutaraldehyde or other fixatives.
Quick Biology scRNA-seq workflow:
We follow a complete WTA workflow from cell lysis, gDNA removal and cDNA synthesis to highly uniform amplification across the entire transcriptome in a one-tube protocol – with negligible sequence bias.

Sample requirements:
· 1–1000 intact cells from all vertebrate species (e.g., human, mouse, rat, sorted cells, tissue culture cells, cells picked under the microscope or microdissected cells from frozen tissue).
· 100 pg – 100 ng of purified RNA
Bioinformatic Service for scRNA-seq:
Standard analysis
b.Alignment to a reference with mapping statistics
d.Differentially Expressed Gene
i. Final project report with analysis methods and results
Advanced analysis
j.Alternative pre-mRNA splicing
n.Fusion genes/transcript detection
Unlock the full potential of your RNA-seq experiment for both lncRNA and mRNA:
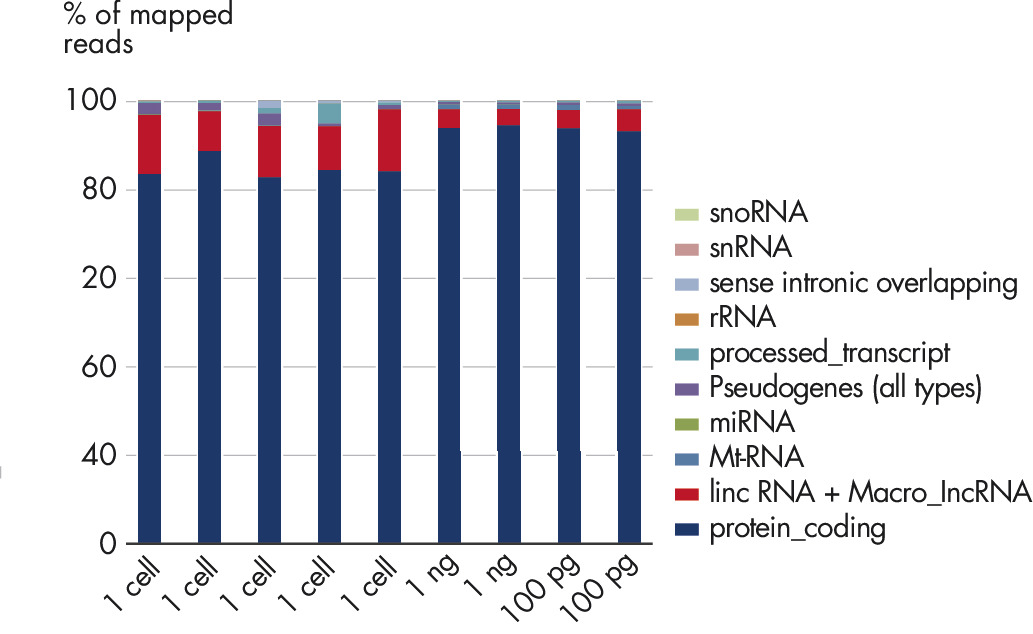
The mapping result shows the number of detected transcripts. Single cell libraries were prepared from PBMCs or total RNA from PBMCs and sequenced on NextSeq. Plotted is the % of reads that map to different RNA biotypes. Besides reads mapping to annotated protein-coding genes, it also unlocks non-coding lncRNA and lincRNA, which can deliver an additional level of insight.
Turnaround time:
About 3-4 weeks, depends on the quality of submitted samples, one additional week for data analysis.

