Quick Biology Inc. has set up 10X Genomics’ Single Cell Fixed RNA Profiling Sequencing platform.
The Single Cell Fixed RNA Profiling (FRP) workflow measures RNA levels in samples (single cells or nuclei) fixed with formaldehyde, using probes targeting the whole transcriptome. After whole transcriptome probe panels are added to the sample, each probe pair hybridizes to its target gene and is then ligated. Libraries are generated from the barcoded probes and sequenced. The final sequencing library structure is similar to Gene Expression solutions for fresh frozen tissue, but the synthetic probe DNA is sequenced rather than the cDNA of a captured transcript.
Key advantages of Fixed RNA Profiling include:
- Process formaldehyde-fixed samples
- Increased sample throughput in a single GEM well with multiplexing
- High-sensitivity WTA profiling with efficient sequencing
Potential to retain multiplet data if each cell has a unique Probe Barcode, illustrated below as a demultiplexible doublet:

Four examples of GEMs with 10x Genomics gel bead (dark blue) and zero, one, or more cells (red or green).
Table of Contents
- Analyzing Fixed RNA Profiling data with Cell Ranger
- Comparing 10x Genomics single-sample solutions
- Comparing 10x Genomics multiplexing solutions
Analyzing Fixed RNA Profiling data with Cell Ranger
The software Cell Ranger v7.0 or later is required to analyze Fixed RNA Profiling data. Cell Ranger v7.1 supports analysis of single cell FFPE (scFFPE) datasets.
One or more samples are each uniquely tagged with a Probe Barcode prior to pooling in a single GEM well, resulting in a Gene Expression (GEX) library for each GEM well. A single-sample ("singleplex") Fixed RNA Profiling experiment with one Probe Barcode and one sample is compatible with Cell Surface Protein Feature Barcode technology, and can generate a paired GEX and Feature Barcode library (hereafter referred to as "Antibody Capture library"). Here are two example workflows:
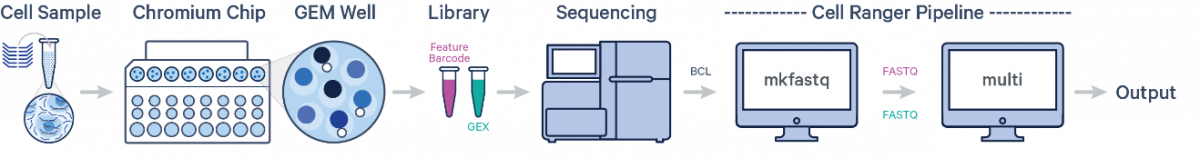
One sample, one Probe Barcode, two libraries:
Multiple samples, multiple Probe Barcodes, one library:
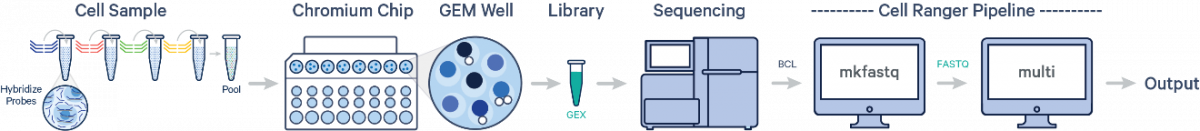
After demultiplexing the BCL files with cellranger mkfastq, run the cellranger multi pipeline on the FASTQ data to obtain separate per-sample output files. Fixed RNA Profiling Gene Expression relies on the same underlying Cell Ranger analysis software as Single Cell 3’ Gene Expression solutions, but uses a short read aligner tailored to probe sequences. The probe aligner assigns probe and gene IDs to reads originating from ligation of correctly paired probe halves, distinguishing from potential artifacts and non-probe reads. For fixed samples, Cell Ranger counts oligo ligation events to build the feature-barcode matrix. As a consequence, if the probe hybridizes to a transcript with a variant, that variant will not be present in the sequencing data. The probe design avoids known SNPs. For information on the probe sets, please visit the Probe Set Overview.
Comparing 10x Genomics single-sample solutions
10x Genomics offers two Chromium products for single-sample gene expression analysis: Single Cell 3' Gene Expression and Single Cell Fixed RNA Profiling for Singleplexed Samples. Key comparisons between these products are summarized below:
|
3' Gene Expression |
Singleplex Fixed RNA Profiling |
|
|---|---|---|
|
Feature Barcode compatibility |
Antibody Capture, CRISPR Guide Capture, 3' Cell Multiplexing |
Antibody Capture (TotalSeq-B) |
|
Capture sequence on gel beads |
Poly(dT) |
Partial capture seq 1 |
|
R2 reads from |
cDNA made from RNA transcripts |
Ligated probes |
|
Cell Ranger pipeline |
count or multi |
multi |
|
R2 reads mapped to |
Reference transcriptome |
Reference probe set |
|
Raw feature-barcode matrix filtered by |
Cell calling |
Cell calling + probe filtering |
Comparing 10x Genomics multiplexing solutions
10x Genomics offers two Chromium products that enable pooling multiple samples in the same GEM well: Single Cell 3' Gene Expression with Cell Multiplexing and Single Cell Fixed RNA Profiling for Multiplexed Samples. Key comparisons between these products are summarized below:
|
3' Cell Multiplexing |
Multiplex Fixed RNA Profiling |
|
|---|---|---|
|
Feature Barcode compatibility |
Antibody Capture, CRISPR Guide Capture |
Not compatible |
|
Sample multiplexing tag |
Lipid-based CMO tag |
Probe Barcode in the probe set |
|
Number of libraries |
Two (GEX and CMO) |
One (GEX) |
|
Cell Ranger pipeline |
multi |
multi |
|
Cell barcode consists of |
10x GEM Barcode + Probe Barcode |
|
|
Tag assignment |
GEM-level |
Read-level |
|
Sample assignment by |
Reads from singlet cell barcodes assigned to this sample |
Reads with a valid barcode (either cell or non-cell) assigned to this sample |
|
Multiplet detection |
Multiplets detected and removed prior to analysis (see true/same-sample doublet diagram above) |
Multiplet data is usable if each cell in the GEM has a unique Probe Barcode (see demultiplexible doublet diagram above) |
|
Cell calling performed on |
The whole GEX library |
Individual samples |
|
Barcode rank plot |
Only one for the whole GEX library |
One for each sample in the pool |
*Citing https://support.10xgenomics.com/single-cell-gene-expression/software/pi…



