2020-10-12 By Quick Biology
Cellular hypoxia (low oxygen) is a stress common in the pathological processes, such as cancers (tumor hypoxia), ischemia (ischemic hypoxia), and preeclampsia (placental hypoxia). It affects cell metabolism, apoptosis, proliferation, and differentiation. Importantly, hypoxia in the Tumor Microenvironment (TME) is the negative factor in the efficacy of radio- and chemotherapy, which becomes a big barrier in cancer treatment.
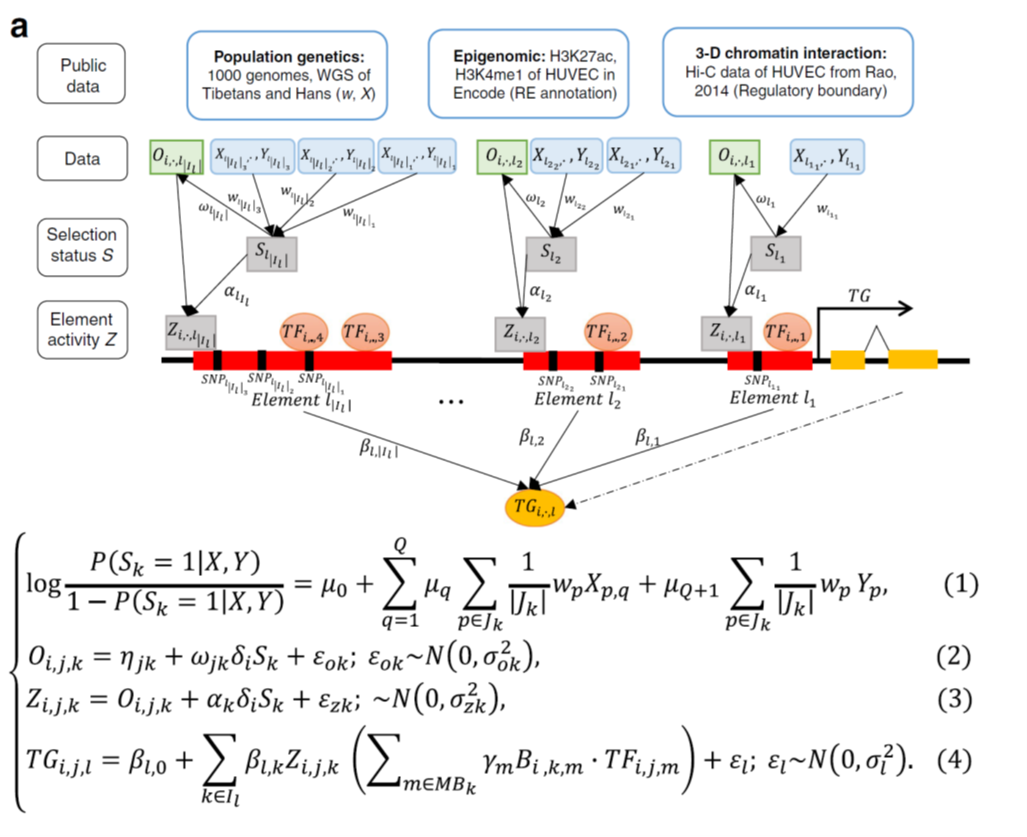
Low oxygen (hypoxia) is also an important stimulus in physiology (e.g. high altitude living). Such a high-altitude adaptation of Tibetans represents a remarkable case of natural selection during recent human evolution. Understanding the genetic basis (such as single nucleotide polymorphisms in Tibetans) and how cells respond to hypoxia and reinstate oxygen homeostasis will enhance our knowledge of hypoxia signal pathways, human adaptive evolution/ecological selection. In recent Nature Communications, evolutionary biologist Prof. Bing Su in State Key Lab of Genetic Resources and Evolution in China, collaborated with biological statistician Prof. Wing Hung Wong at Stanford, Prof. Yong Wang in the Academy of Mathematics and Systems Sciences in Beijing, performed time-courses of paired ATAC-seq, RNA-seq on collected HUVECs (human umbilical vein endothelial cells) with different genotypes under hypoxia and normoxic conditions (Ref1. Fig.1). By integrating population data of Tibetans and Hans, Hi-C data of HUVEC cells, researchers further developed method called vPECA (variant interpretation methodology, Fig.2), discovered that three SNPs of EPAS1 can decrease the accessibility of active selected non-coding regulatory elements (ASREs), contribute to hypoxia adaptation. This work provides us a systematic approach to interpret phenotype-associated noncoding variants in cells.
Figure 1: Experimental design for time-series hypoxia induction and multi-level omics data profiling. (ref1)
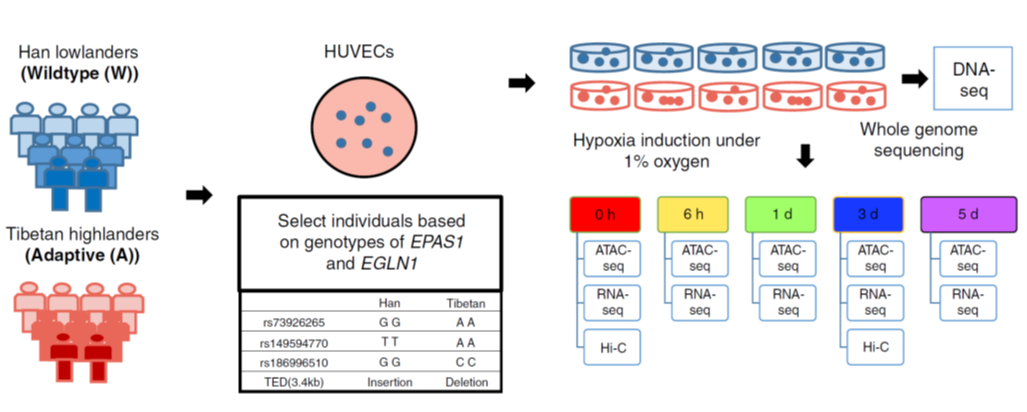
Figure 2: vPECA models how positively selected noncoding SNPs affect the RE’s selection status, chromatin accessibility, and activity, and further determine the target gene’s (TG) expression by integrating paired expression and chromatin accessibility profiling with population genetics, epigenome, and 3D chromatin interaction data. (ref1)
Quick Biology provides RNA-seq, ATAC-seq, and Hi-C services. Find More at Quick Biology.
Ref:
1. Xin, J. et al. Chromatin accessibility landscape and regulatory network of high-altitude hypoxia adaptation. Nat. Commun. 11, 4928 (2020).



