Introduction
100-QuickPathTM siRNA Arrays
Quickly Study Gene-Gene, Gene-Drug interactions in 100 Pathways with Cell-Based Assays
The 100-QuickPathTM siRNA Array is the latest development by Quick Biology Inc. and is a great tool for conducting functional studies on your gene of interest through siRNA treated cell-based assays. Validated siRNA for key pathway-focused genes with appropriate controls are arrayed on each 96-well plate. Following a simple cell culture protocol with the siRNA in the plate, you can quickly and directly determine gene-gene or gene-drug interactions on the same plate with colorimetric, luminescent, or fluorescent cell-based detection methods. In the current version, the targets of the selected siRNAs include drug targets, significantly mutated genes (cancer driver genes) and transcription factors. The goal of this siRNA Array set is to use siRNAs as tools to survey potential gene-gene or gene-drug interactions.
Order Information:
|
Product |
Drug Targets (click to check targeted genes) |
Cat. #. |
Unit Price (USD) |
|
QV0201 |
$289 per array
|
||
|
QV0202 |
|||
|
QV0203 |
* The cancer driver genes are selected based on a Nature paper: "Discovery and saturation analysis of cancer genes across 21 tumour types, Nature 505,495–501, 2014".
![]() QB siRNA Array User Manual v4.pdf
QB siRNA Array User Manual v4.pdf
![]() siRNA Array data analysis template v4.xlsx
siRNA Array data analysis template v4.xlsx
Please click the corresponding siRNA array product to see the detailed plate design and gene targets.
Why Use siRNA Array?
l Quickly obtained biological insights by perturbing the expression of a panel of genes related to a pathway or disease state.
l Serves as a good alternative to Drug Array in screening for potential gene-drug interactions. .
l siRNAs are usually more specific than most selective chemicals for perturbing gene expressions.
l siRNA can target non-druggable genes which are critical in cancer and other disease.
l Easily applied to high throughput and routine use in any biomedical research labs.
100-QuickPathTM siRNA Array Applications:
In the current version, the targets of the selected siRNAs include drug targets, significantly mutated genes and transcription factors highly relevant to cancer. One potential application for using siRNA Arrays is highlighted in the following use case.
--Screening genes regulating anti-cancer drug resistance.
Depletion of the targeted genes therefore sensitizes the cancer cells to anti-cancer drugs, sometimes leading to synthetic lethality. Inducing synthetic lethality in cancer cells is indeed a valuable strategy to selectively kill cancer cells with minimal effect on normal cells.
Example 1: TP53 is an important player in DNA damage signaling pathways. When TP53 is mutated in some tumors, other G2/M checkpoint regulators such as checkpoint kinase 1 (CHK1) compensates for the absence of TP53 and arrests the cell cycle. If both TP53 and CHK1 are inhibited, the tumor cells are unable to arrest cell cycle for DNA damage repair, and thus the accumulation of unrepaired DNA will reduce the viability of tumor cells [1].
Example 2: The myelocytomatosis viral oncogene homolog (MYC) is a transcription factor that plays an important role in cell cycle progression, apoptosis, and cellular transformation. Death Receptor 5 (DR5), a member of the TNF-receptor superfamily, is a mediator of apoptosis. MYC is over expressed in some cancers, and it has been shown in fibroblasts that increased expression of MYC causes sensitivity to agonists of DR5 [2].
How it works?
--Simple Experimental Designs and Workflow:
Each siRNA Array plate is designed to compare two different samples in parallel on one ready-to-use 96-well plate. The siRNAs are supplied in a lyophilized form, and each well contains 2.5 pmol of siRNA (final concentration will be 25nM after adding 100 μl of transfection mixture and cells. Following a simple cell culture protocol with the siRNA in the plate, you can quickly and directly determine the phenotype of cells on the same plate with colorimetric, luminescent, or fluorescent cell-based detection methods. Please find siRNA Array product manual here.
![]() QB siRNA Array User Manual v4.pdf
QB siRNA Array User Manual v4.pdf
1. Drug-gene interaction
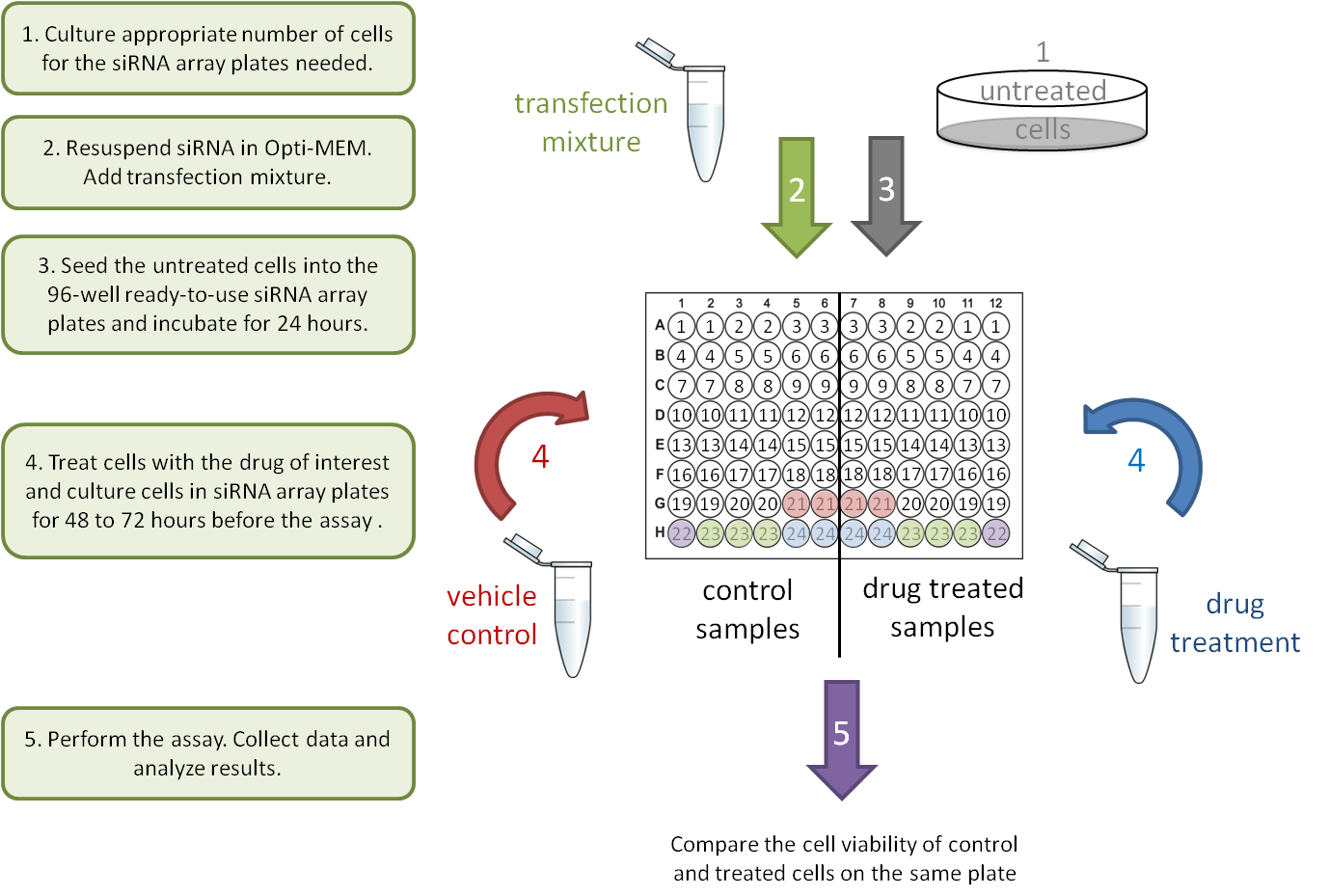
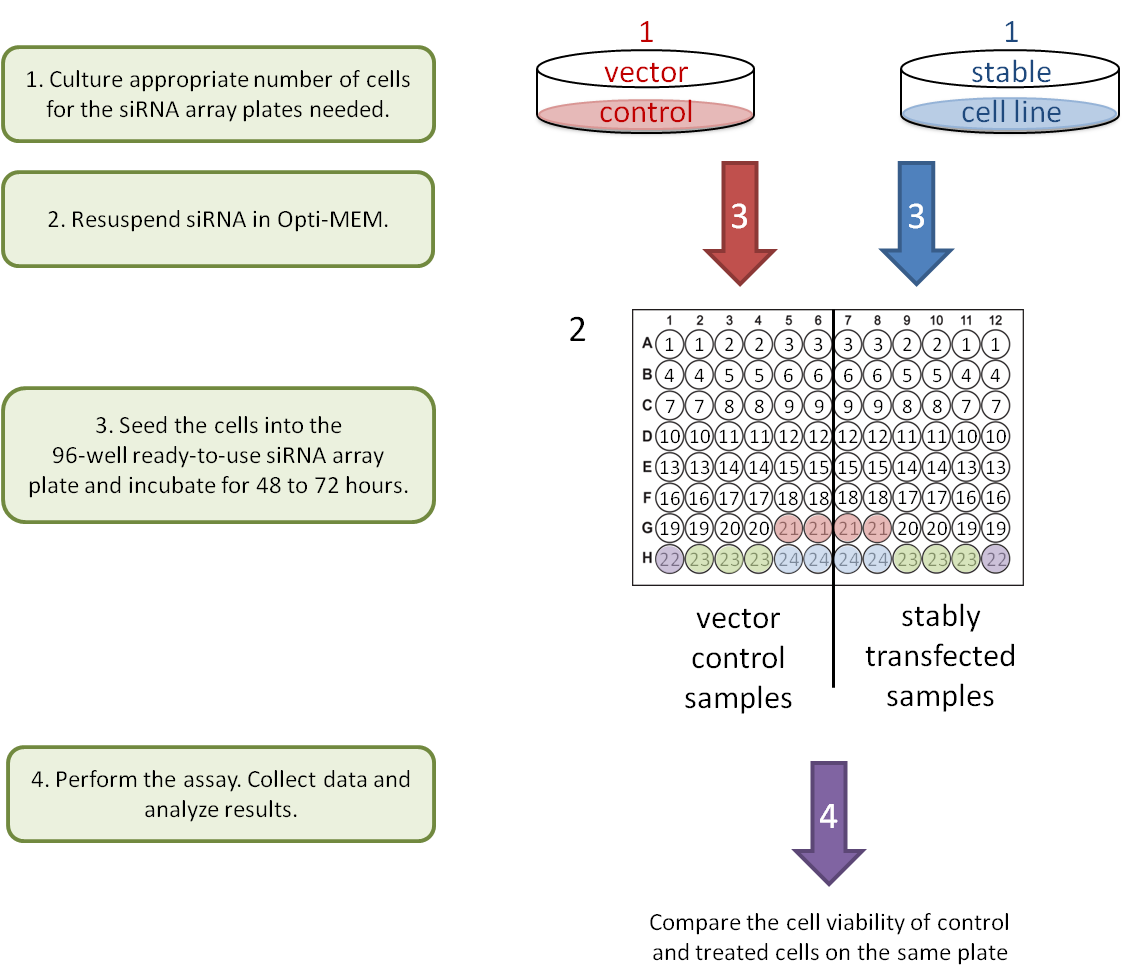
2. Gene-Gene interaction: stable knockdown/over-expression cell lines
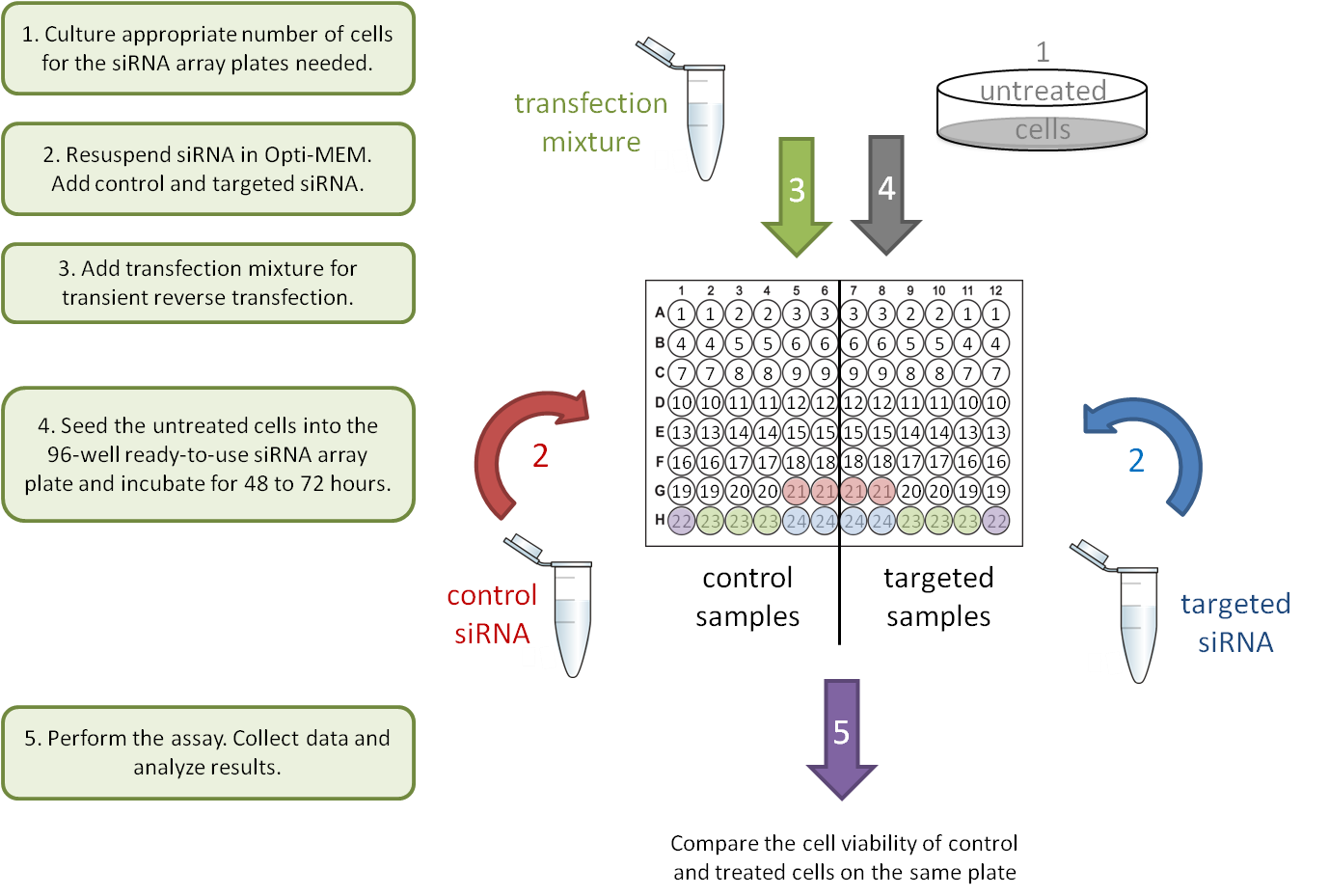
3. Gene-Gene interaction: transient knockdown
--Ready-to-use data analysis:
Quick Biology presents a user-friendly analysis template in excel format, which can be downloaded from here.
![]() siRNA Array data analysis template v4.xlsx
siRNA Array data analysis template v4.xlsx
The users will only need to input their own data into the Raw Data sheet. The pre-programmed algorithm on the Analysis sheet will subtract the background (plate well # 22), normalize against the scrambled siRNA control (plate well # 23), and calculate the average, standard deviation, and log2 ratio for each treatment and control pair. The results will be transformed into figures for easy interpretations.
Reference:
(1) Morandell, S. and M.B. Yaffe, Exploiting synthetic lethal interactions between DNA damage signaling, checkpoint control, and p53 for targeted cancer therapy. Prog Mol Biol Transl Sci, 2012. 110: p. 289-314.
(2) Wang, Y., et al., Synthetic lethal targeting of MYC by activation of the DR5 death receptor pathway. Cancer Cell, 2004. 5(5): p. 501-12.



