2020-02-16 by Quick Biology Inc.
RNA plays a special role in life science. It can be as genetic materials like DNA, it can catalyze specific biochemical reactions, which is similar to the action of the protein. Due to its single strand, it forms many secondary structures, providing more variation ------ a significant evolutionary advantage compared to DNA. One specific RNA type whenever is discovered will become a fascinating research topic, such as miRNA, piRNAs, non-coding RNAs, circular RNAs. Today, we introduce cp-RNA, an RNA harboring a 2’, 3’-cyclic phosphate at 3’-end (Fig.1). Usually, these short RNA molecules are generated by ribonuclease cleavage, which seems like non-functional degradation products. Due to specific 3’ end, those cP-RNAs are not ligated to a 3’-adapter and thus uncaptured by standard RNA-seq. In PLOS Genetics, Kirino Lab developed a novel method (Fig.2, ref 1) for large screening cP-RNA molecules in the transcriptome (Fig.1, ref 1). By performing cP-RNA-seq across multiple mouse tissues, they characterized cP-RNA molecules where and how they are generated, they also found these cP-RNAs were derived from specific sites of substrate RNAs, the expression levels showed age-dependent reduction (Fig.3-4, ref2). These patterns strongly indicate that cp-RNA may have physiological roles in various biological processes, not just accumulated as a degradation product.
Figure 1: 3’-end Structure of regular RNA and cP-RNAs, and reactivity with enzymatic treatments. T4 PNK removes both a 3′-P and 3′-cP to form a 3′-OH end. A phosphatase such as CIP removes a 3′-P (and 5′-P) but not a 3′-cP. An HCl treatment followed by CIP treatment (HCl + CIP) can remove a 3′-cP because the HCl treatment converts 3′-cP to a 3′-P (ref 1).
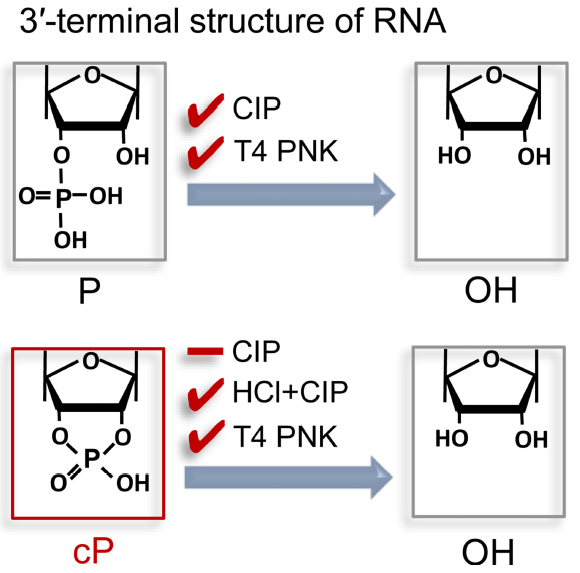
Figure 2: A schematic representation of the cP-RNA-seq procedure for selective amplification and sequencing of cP-containing RNAs. Total RNA is extracted from cell lines or tissues (Steps 1–10), and then the RNAs with targeted lengths are gel-purified using denaturing PAGE (Steps 11–27). The purified RNAs are subjected to CIP treatment (Steps 28–36), periodate treatment (Steps 37–40), and T4 PNK treatment (Steps 41–43). By using an Illumina kit, the treated RNAs are ligated to adapters (Steps 44–53) and their cDNAs are amplified by RT-PCR (Steps 54–59). The amplified cDNAs are gel-purified using native PAGE (Steps 60–72), and then subjected to Illumina next-generation sequencing. (ref2)
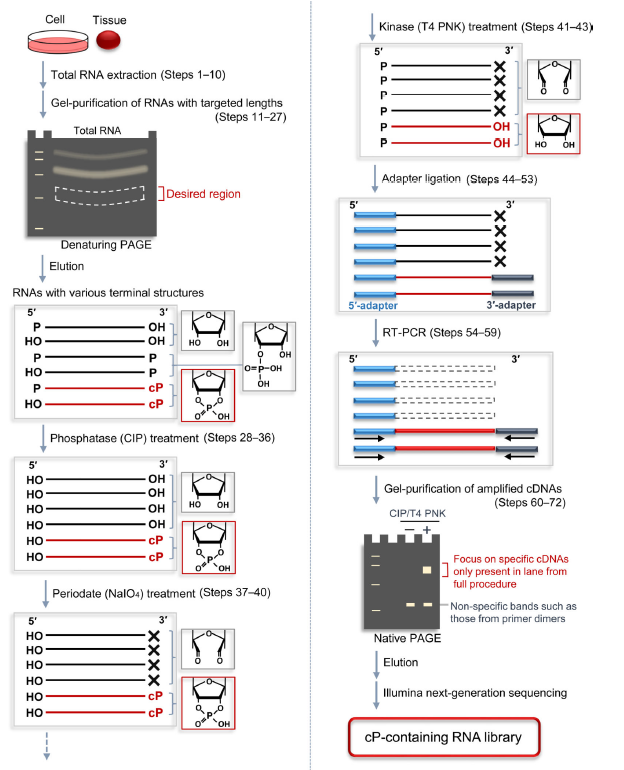
Figure 3: Predicted cleavage sites in anticodon-loops in thymus. Cleavage sites in the anticodon-loops in the thymus were predicted based on the 30-terminal positions of the 50-halves (blue arrow heads) and the 50-terminal positions of the 30-halves (red arrow heads) (ref2).
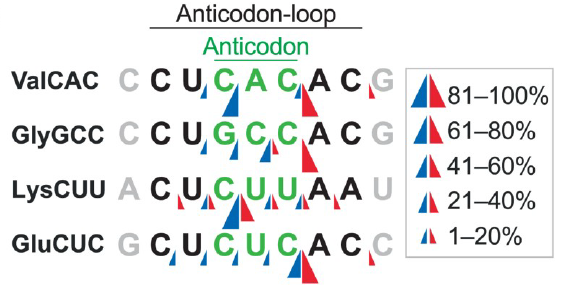
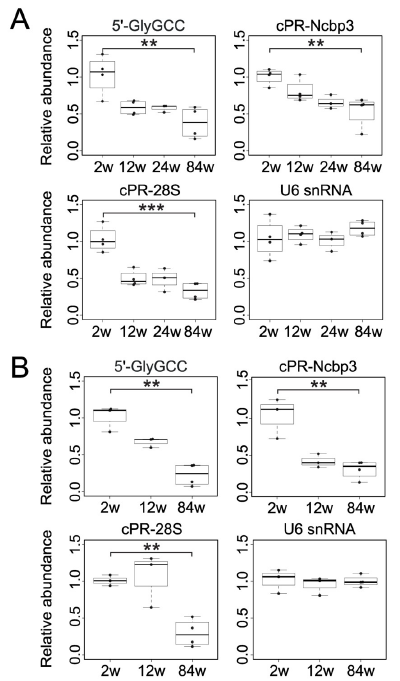
 Figure 4: Expression of cP-RNAs in skeletal muscle (A) and lung from mice with different ages. (A) Total RNAs from skeletal muscle of 2–84-week old mice were subjected to TaqMan RT-qPCR for quantification of the indicated cP-RNAs. U6 snRNA was quantified as the control whose expression is unchanged through aging.P < 0.01, P < 0.001 by t-test. (B) Total RNAs from lung of 2–84-week old mice were subjected to cP-RNA quantification (ref2).
Figure 4: Expression of cP-RNAs in skeletal muscle (A) and lung from mice with different ages. (A) Total RNAs from skeletal muscle of 2–84-week old mice were subjected to TaqMan RT-qPCR for quantification of the indicated cP-RNAs. U6 snRNA was quantified as the control whose expression is unchanged through aging.P < 0.01, P < 0.001 by t-test. (B) Total RNAs from lung of 2–84-week old mice were subjected to cP-RNA quantification (ref2).
Quick Biology is expert and interested in novel RNA-seq development. Find More at Quick Biology.
Ref:
1. Honda S, Morichika K, Kirino Y. Selective amplification and sequencing of cyclic phosphate-containing RNAs by the cP-RNA-seq method. Nat Protoc. 11, 476–489 (2016).
2. Shigematsu, M., Morichika, K., Kawamura, T., Honda, S. & Kirino, Y. Genome-wide identification of short 20,30-cyclic phosphate-containing RNAs and their regulation in aging. PLoS Genet. 15, 1–15 (2019).



