4/12/2019 by Quick Biology Inc.
Next-generation sequencing (NGS), also known as high-throughput sequencing technologies allow us to sequence DNA and RNA much more quickly and cheaply than the previously used Sanger sequencing, and as such have revolutionized the study of genomics and molecular biology.
Within the last 2–3 years, we have witnessed a rapid increase in quantity and quality in genomic and transcriptomic research that expands into other “-omics” fields. Technological changes allowed the expansion of our knowledge and changed views on bacterial genetics and biology. In this crash course in next-generation sequencing (NGS) for a microbiologist, researchers from National Medicines Institute in Warszawa, Poland discuss and review the applications of high-throughput Next-generation sequencing (NGS) methods to study bacteria in a much broader context than simply their genomes. Today, the major use of next-generation sequencing including: study the complex regulatory processes in the bacterial cells; and study the organization of the bacterial life—from complex communities to single cells, extending its capabilities to diagnostic microbiology and epidemiology.
Key Points:
1. Affordable high-throughput methods on modern microbiology change of the way we think about the experimental approach.
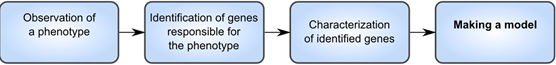
Figure 1. Classical methods lead from single observations of biological phenomena to generating a model of activity.
Now, we can use multiple high-throughput NGS methods, including genomic studies, combined with data mining, allow drawing a broader complete picture (Figure 2).
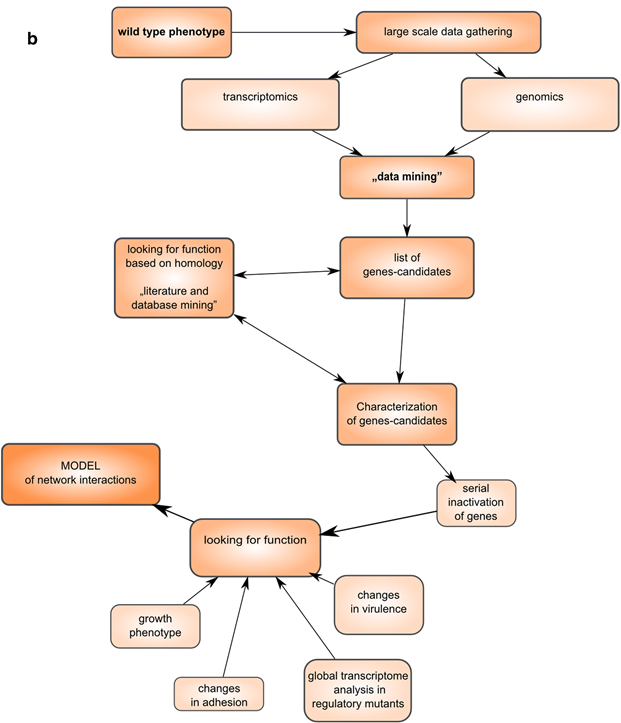
2. Transcriptomics and microbiome analysis based on a combination of multiple levels of the “-omics” data.
Today, most of transcriptomics techniques (SAGE, quantitative PCR and later microarrays) are currently being replaced by RNA sequencing technologies. Combining It allows to expand studies on basic bacterial biology such as transcription, translation, and the interaction of basic molecules in the living cells (Fig. 3) and link multiple levels of information such as genomic and transcriptomic data. NGS is great tool for a complex analysis of microbial communities by quantitatively assessing the composition and proportions of species within various microbiomes. This type of sequencing are ITS and/or 16s/18S rRNA sequencing. What’s more, metagenome sequencing (WGS) enable us to study the metabolic properties of the bacterial community or co-evolution of the host and bacteria and also allows to characterize unique bacterial communities and bacteria. This type of sequencing prompted the rapid development of microbiome meta-transcriptomics, especially for the microbiota and gut interactions. As a result, we can now combine multiple level of NGS data, such as alternate transcripts, non-coding RNAs, overlapping UTRs, leaderless mRNA, riboswitches, antisense RNA, regulation by genome structure, or epigenetic modifications (Figure 3).
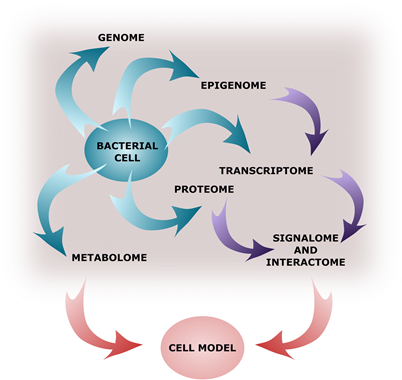
Figure 3. A virtual cell reconstruction that will be possible in the future based on a combination of multiple levels of the “-omics” data
Recently, as 10X Genomics gaining more popular, its single cell WGS and RNA-seq allow us to sequence material isolated from single cells enabled the genome assembly of new phyla and studying transcriptome on the single-cell level, thus provide new biological insights into the microbial world.
3. The major challenge in understanding genomic and transcriptomic data lies today in combining it with other sources of data such as proteome and metabolome.
If you are interested, Quick Biology has the complete solution from sample to insightful data for your microbiology project using RNA-seq, metagenomics, ITS/16s/18S rRNA sequencing and single cell sequencing.
Source: Kozińska A, Seweryn P, Sitkiewicz I. (2019) A crash course in sequencing for a microbiologist.J Appl Genet [Epub ahead of print]. [article]



