2019-10-06 by Quick Biology Inc.
Modification to RNA species (Fig 1) has been discovered for over 50 years. The internal modification m6A N6-methyladenosine (m6A) is the most abundant type, plays diverse roles in a variety of physiological processes.
To date, most global m6A detection relies on RNA immunoprecipitation by m6A-recognizing antibodies, such as MeRIP-seq, m6A-seq. These methods are time-consuming and costly (expensive antibody, a large amount of input RNA). In recently Nature Methods, Dr. Meyer developed DART-seq: an antibody-free method for global m6A detection. In this experiment, Dr. Meyer fused APOBEC1 to the m6A-binding YTH domain. Recruitment of APOBEC1 to m6A site will enable deamination of cytidine immediately following m6A residue like C-to-U editing. APOBEC1-YTH is transiently expressed in cells and total RNA is isolated and subjected to RNA-seq. C-to-U mutations are then detected to identify sites of m6A (Fig 2).
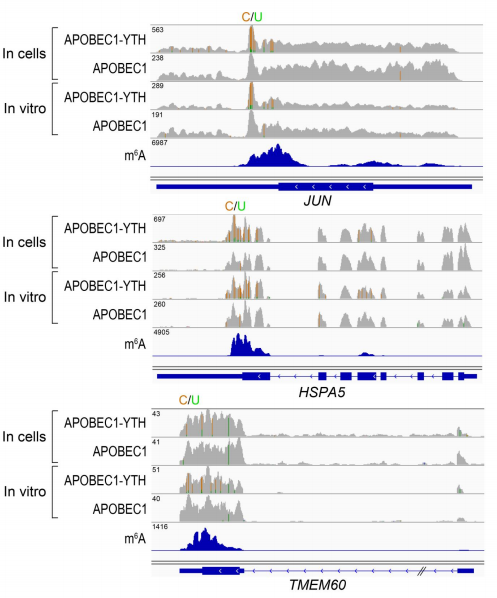
To validate DARP-seq approach, the author performed DART-seq, compared to m6A sites discovered previously by miCLIP (Fig 3). To rule out any potential cell alteration caused by APOBEC1-YTH transfection in cell, she also performed in vitro DART-seq just using HEK293T cell RNA treated by APOBEC1-YTH protein (Fig 4). Both data show great agreement with traditional antibody-based methods.
Figure 1: Chemical modification in eukaryotic messenger RNAs (ref1). A schematic representation of common chemical modifications in eukaryotic mRNA transcripts. Several of these modifications map uniquely to the mRNA cap structure, 5’ or 3’ untranslated regions, or the coding region (bold) of the transcript.
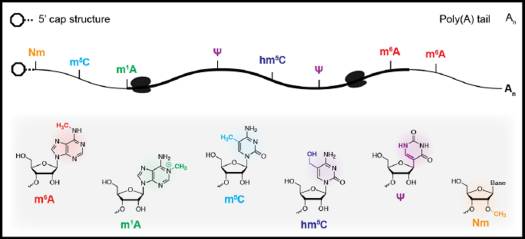
Figure 2: Schematic of DART-seq (ref2). APOBEC1 is fused to the YTH domain to guide C-to-U editing at cytidine residues adjacent to m6A sites. APOBEC1-YTH is expressed in cells and total RNA is isolated and subjected to RNA-seq. C-to-U mutations are then detected to identify sites of m6A.
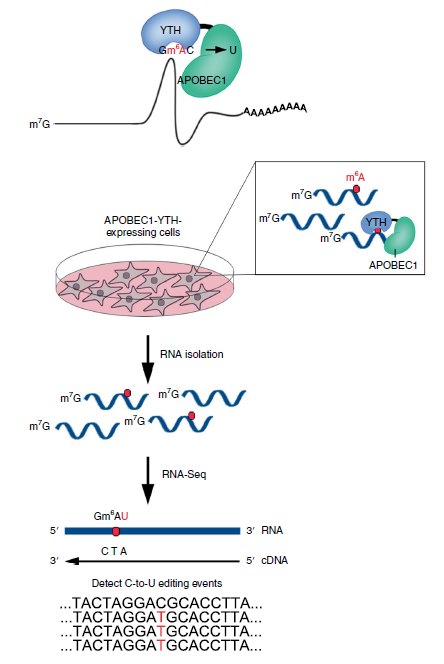
Figure 3: Example of m6A sites detected by DART-seq (ref2). C-to-U mutations found in at least 10% of reads are indicated by gold/green coloring at individual sites (gold indicates the abundance of C sites and green indicates the abundance of U sites at each position; ACTB is transcribed from the negative strand). Percentages of C-to-U mutations at two individual sites previously identified by miCLIP5 are indicated in the expanded view. Results are representative of three independent experiments.
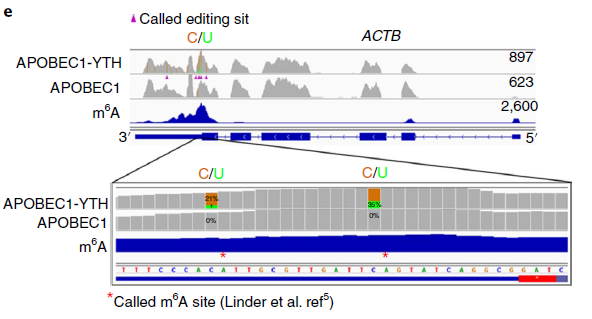
Figure 4: In Vitro DART-seq identifies m6A sites in cellular RNAs. (ref2)
Quick Biology is an expert in Next Generation Sequencing. Are you ready to sequence your samples of interest? Find More at Quick Biology.
Reference:
- 1. Roundtree etc Dynamic RNA modifications in gene expression regulation. Cell. 169, 1187–1200
- 2. Meyer, K. D. DART-seq: an antibody-free method for global m6A detection. Nat. Methods (2019). doi:10.1038/s41592-019-0570-0 (2017).



