2020-08-10 by Quick Biology Inc.
N6-methyladenosine (m6A) is the most abundant modification of mRNA. This modification has critical roles in stem cell proliferation, differentiation, mRNA splicing, RNA metabolism, etc. Previously, we described the DART-seq method developed by Dr. Kate D.Meyer, an antibody-free method for global m6A detection. In DART-seq, the researcher fused APOBEC1 to the m6A-binding YTH domain. Recruitment of APOBEC1 to m6A site will enable the deamination of cytidine immediately following m6A residue like C-to-U editing.
Today, we introduce the other three antibody-independent methods developed by researches in China:
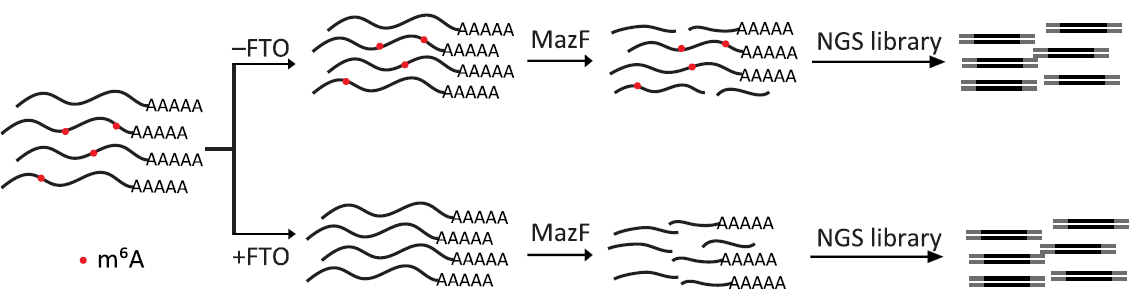
1. m6A-REF-seq (m6A-sensitive RNA-Endoribonuclease-Facilitated sequencing) by Dr. Luo at Sun Yat-sen University. (2019 July Sci.Adv). This method is based on the m6A-sensitive RNA endoribonuclease (i.e MazF) recognizing ACA motif. In general, mRNA is digested into fragments by MazF, fragments are ligated to NGS adaptors. To prevent false positives, a parallel sample can be demethylated by fat mass and obesity-associated protein (FTO) (Fig.1).
Figure 1: Schematic diagram of m6A-REF-seq. fat mass and obesity-associated protein (FTO), source from ref 1.
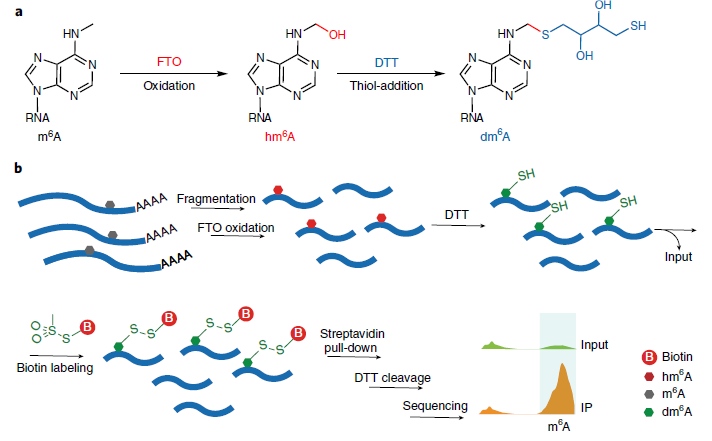
2. m6A-SEAL (FTO-assisted m6A selective chemical labeling method) by Dr. Jia at Peking University. (2020 Aug. Nature Chemical Biology). In this method, FTO oxidatation of m6A and a DTT-mediated thiol-addition reaction will convert m6A to dm6A. Followed by Biotin labeling and streptavidin pull-down and DTT cleavage, it can enrich m6A sites in the whole transcriptome (Fig.2).
Figure 2: Schematic diagram of m6A-SEAL (ref2). a, FTO oxidation of m6A to hm6A and a DTT-mediated thiol-addition reaction to convert unstable hm6A to the more stable dm6A. b, Schematic illustration of the m6A-SEAL-seq pipeline.
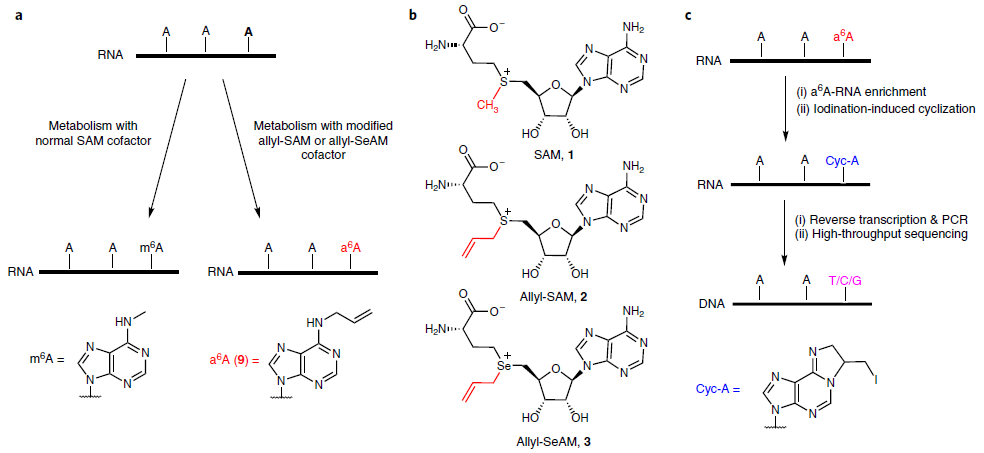
3. m6A-label-seq (a metabolic labeling m6A method) by Dr. Liu at Zhejiang University. (2020 Aug. Nature Chemical Biology). In this method, cells are fed with a methionine analog, Se-allyl-l-selenohomocysteine, which substitutes the methyl group on the enzyme cofactor SAM with the allyl. Cellular RNAs could, therefore, be metabolically modified with N6-allyladenosine (a6A) at supposed m6A-generating adenosine sites (Fig.3).
Figure 3: Schematic diagram of m6A-label-seq (ref3). a, Schematic of metabolic labeling of the supposed m6A site with a6A using an allyl-SAM (SeAM) cofactor. b, The chemical structures of SAM, allyl-SAM and allyl-SeAM. c, A simplified procedure to locate metabolically labeled a6A, transcriptome-wide, at single-base resolution.
Quick Biology is an expert in implementing NGS assay. Find More at Quick Biology.
Ref:
- 1. Zhang, Z. et al. Single-base mapping of m 6 A by an antibody-independent method. Science Advances (2019;5:eaax0250)
- 2. Wang, Y., Xiao, Y., Dong, S., Yu, Q. & Jia, G. Antibody-free enzyme-assisted chemical approach for detection of N 6-methyladenosine. Nat. Chem. Biol. 16, 896–903 (2020).
- 3. Shu, X. et al. A metabolic labeling method detects m6A transcriptome-wide at single base resolution. Nat. Chem. Biol. 16, 887–895 (2020).



