2021-04-15 By Quick Biology
Chromatin packing and histone modification are important factors for gene regulation. ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing), and chromatin immunoprecipitation (ChIP)-seq are typical methods for chromatin accessibility and histone modifications. While the genomic profiles of various chromatin and histone modifications have been extensively characterized at the bulk cells, biotech is now at the stage of single-cell resolution.
In recent Genome Research, a team from Dr. Keji Zhao, an epigenetic scientist, developed indexing single-cell immunocleavage sequencing (iscChIC-seq), a multiplex indexing method based on TdT terminal transferase and T4 DNA ligase mediated barcoding strategy and single-cell ChIC-seq, which is capable of readily analyzing histone modifications across tens of thousands of single cells in one experiment (ref1).
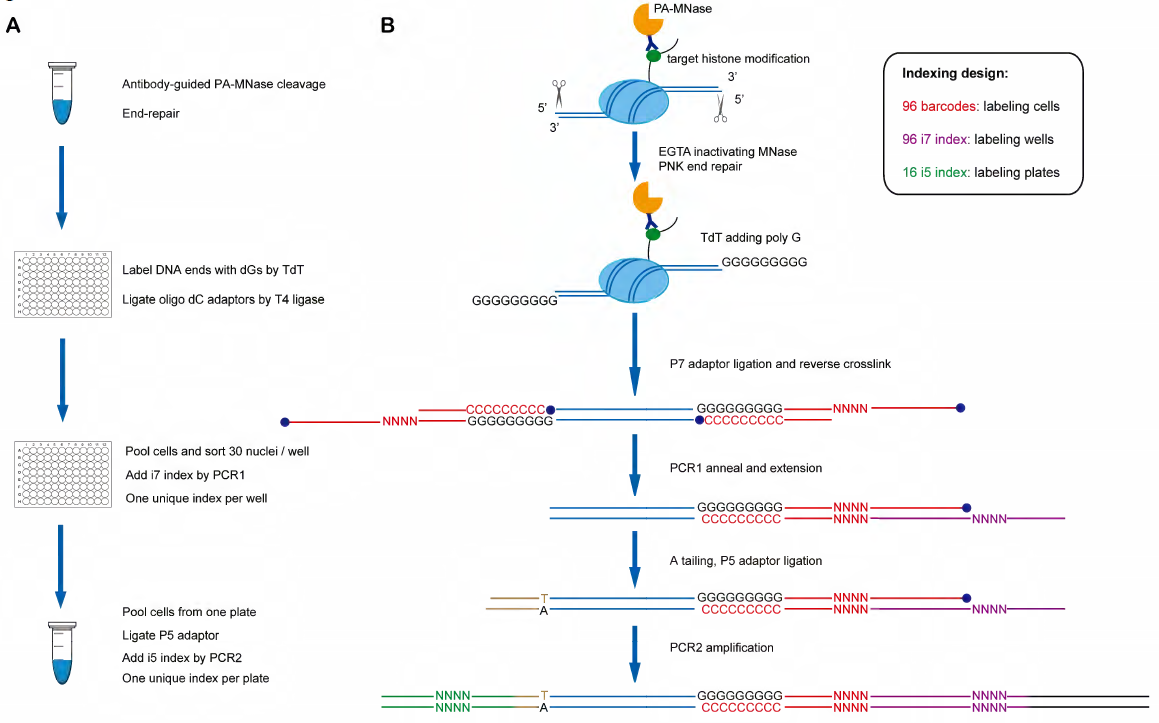
Basically, (see Figure. 1) the procedure is the following:
- a. bulk cells were split into the first 96-well plate after antibody-guided MNase cleavage and end repair.
- b. Poly(G) was added to DNA ends by TdT, oligo dC adaptor ligation by T4 DNA ligase.
- c. After reverse crosslinking, barcoded DNA fragments could be efficiently labeled with i7 index (purple) through annealing and PCR extension.
- d. The barcoded P5 adaptor is added to the other end of genomic DNA fragments by ligation and PCR2, which is used to amplify the library DNA for NGS sequencing.
Using iscChIC-seq to profile H3K4me3 and H3K27me3 in human white blood cells (WBCs) enabled successful detection of more than 10,000 single cells for each histone modification with 11K and 45K non-redundant reads per cell. These data indicate iscChIC-seq is a reliable technique for profiling histone modifications in a large number of single cells.
Figure 1: Experimental flow and illustration of adapter, barcode addition to fragments. (ref1)
Quick Biology can assist you with any NGS-based projects. Find out More at Quick Biology.
See resource:
1. Ku, W. L., Pan, L., Cao, Y., Gao, W. & Zhao, K. Profiling single-cell histone modifications using indexing chromatin immunocleavage sequencing.



