Gastrointestinal adenocarcinomas (GIAC) of the tubular gastrointestinal (GI) tract including esophagus, stomach, colon, and rectum comprise most GI cancers and share a spectrum of genomic features. However, the unified epigenomic changes specific to GIAC are poorly characterized. Using 907 GIAC samples from The Cancer Genome Atlas, we applied mathematical algorithms to large-scale DNA methylome and transcriptome profiles to reconstruct transcription factor (TF) networks and identify a list of functionally hyperactive master regulator (MR) TF shared across different GIAC. The top candidate HNF4A exhibited prominent genomic and epigenomic activation in a GIAC-specific manner. A complex interplay between the HNF4A promoter and three distal enhancer elements was coordinated by GIAC-specific MRTF including ELF3, GATA4, GATA6, and KLF5. HNF4A also self-regulated its own promoter and enhancers. Functionally, HNF4A promoted cancer proliferation and survival by transcriptional activation of many downstream targets, including HNF1A and factors of interleukin signaling, in a lineage-specific manner. Overall, this study provides new insights into the GIAC-specific gene regulatory networks and identifies potential therapeutic strategies against these common cancers.
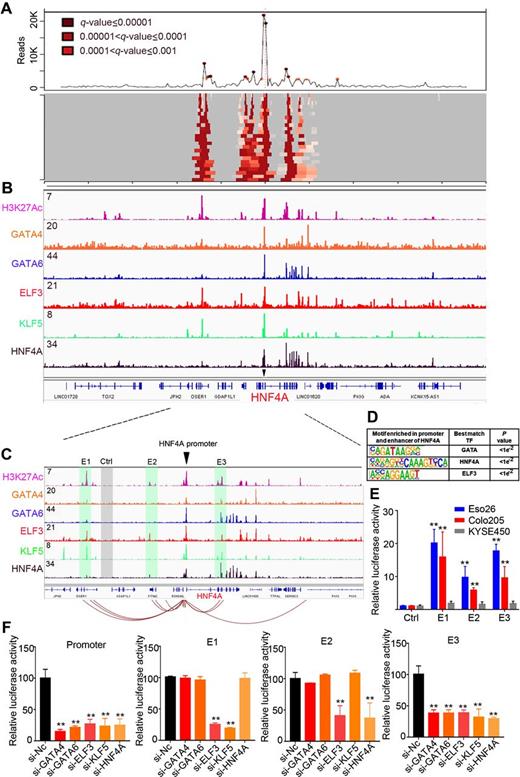
n the Figure, GIAC MRTFs occupy and regulate HNF4A promoter and enhancers. A, 4C-Seq assay showing the long-range interactions anchored on HNF4A promoter in Eso26 cells. Deeper red color indicates higher interaction frequency. B, ChIP-seq profiles for H3K27Ac and indicated MRTFs at HNF4A loci in Eso26 cells. C, Zoom in view of ChIP-seq signals. Connecting lines showing the most significant interactions detected by 4C. Shaded regions showing three enhancers (E1–E3) and one negative control (Ctrl) region that were separately cloned into the luciferase reporter vector. D, Motif enrichment analysis of promoter and E1–E3 regions. E, Enhancer activity measured by luciferase assays in Eso26, Colo205, and KYSE450 cells. F, Enhancer and promoter activity measured by luciferase assays in Eso26 cells upon silencing each of the 5 MRTFs. Mean ± SD are shown. **, P < 0.01.
Quick Biology provided the service of the CHIP sequencing for this study.
ChIP-sequencing, also known as ChIP-Seq or ChIP-seq, is a method used to analyze protein interactions with DNA. It combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. ChIP-seq is used primarily to determine how transcription factors and other chromatin-associated proteins influence phenotype-affecting mechanisms. Determining how proteins interact with DNA to regulate gene expression is essential for fully understanding many biological processes and disease states. This epigenetic information is complementary to genotype and expression analysis.
Quick Biology is a professional ChIP-Seq provider and will ensure the customer to get the most out of the sequencing project.
See resource:
1. Pan, J., Silva, T. C., Gull, N., Yang, Q., Plummer, J. T., Chen, S., ... & Lin, D. C. (2020). Lineage-specific epigenomic and genomic activation of oncogene HNF4A promotes gastrointestinal adenocarcinomas. Cancer research, 80(13), 2722-2736.



