2020-02-23 by Quick Biology Inc.
Extrachromosomal DNA (ecDNA) is any DNA that is found off the chromosomes. Typically, for instance in human cell, mitochondrial DNA (mtDNA) is one type of ecDNA. Today we are going to talking about another type---extrachromosomal circular DNA (eccDNA), which is derived directly from genomic DNA. The function of eccDNA has not been well studied, however, eccDNA is commonly observed in human cancer cells. In Nature, researchers in UCSD and Stanford integrated ultrastructural imaging, long-range optical mapping to characterize the structure of eccDNA in human cancer lines (Fig1-2). By RNA-seq, they revealed that oncogenes encoded on eccDNA are the most highly expressed genes in tumors (Fig3). By ChIP-seq, MNase-seq, and ATAC-seq, they proved eccDNA lacks the higher-order compaction compared to chromosomal DNAs, it is accessible DNA in cancer genomes (Fig4). By PLAC-seq and 4C-seq (long-range chromatin interaction sequencing), researchers reveal eccDNA enables distal DNA interaction (Fig5). Taken together, all these data provide insight into how the structure of eccRNA affects oncogenesis.
Figure 1: Global workflow to characterize the structure and function of ecDNA. (ref1)
Figure 2: Correlated SEM and confocal light microscopy of chromosomal and ecDNA in COLO320DM cell. FISH validating MYC-containing ecDNA in COLO320DM cells visualized by 3D-SIM (ref1).
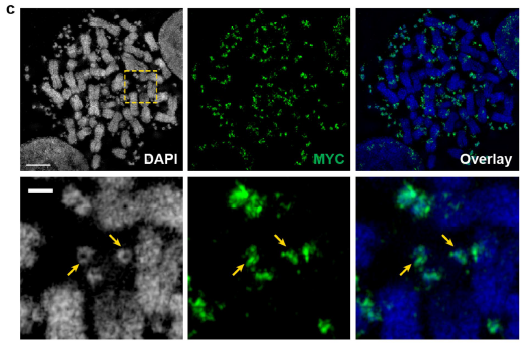
Figure 3: Panel f: Gene copy number comparing circular and linear amplifications (8,068 circular and 6,247 linear amplified (AMP) genes from 77 samples). P-value determined by a two-sided Wilcoxon test. Panel g, Depiction of the mechanism of massive transcript levels from ecDNA. (ref1)
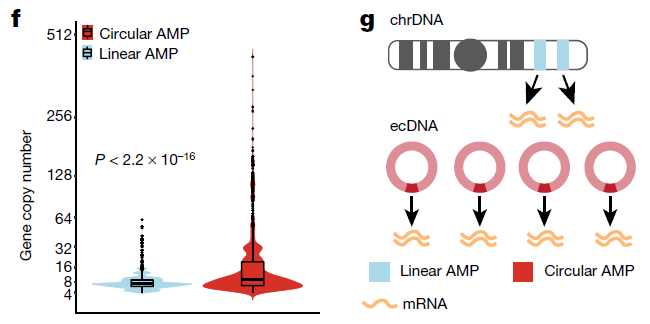
Figure 4: A model depiction to assess chromatin compaction (ref1). ATAC-see identifies unanticipated MYC ecDNAs in GBM39 cells because of their high signal, which was subsequently confirmed by ATAC-seq and WGS.
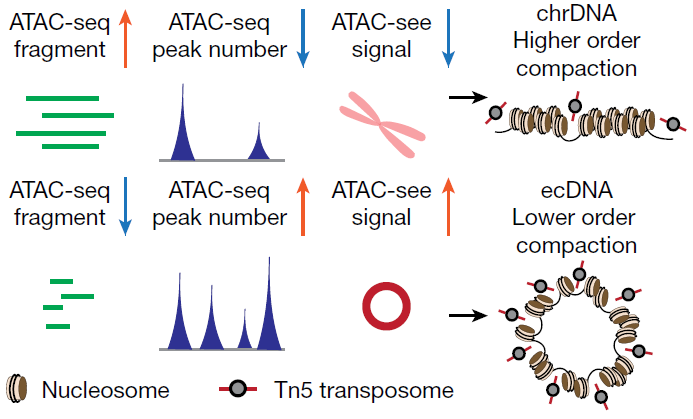
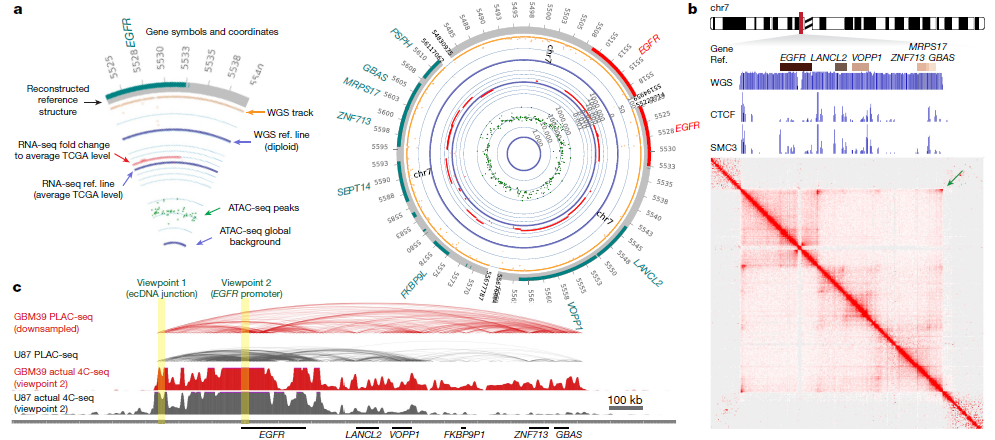
Figure 5: Circularization of ecDNA enables distal DNA interaction. a, Circular plot of ecDNA structure in GBM39 cells. Plot legend is shown on the left. b, MAGIC CollaborationH3K27ac anchored active chromatin interaction heatmap by PLAC-seq and HiChIP in GBM39 cells. WGS depicts the ecDNA amplicon. ChIP–seq demonstrates CTCF and SMC3 binding to ecDNA. Arrow indicates the increased corner reads in ecDNA junction. c, Composite view of PLAC-seq, HiChIP and actual 4C-seq. Virtual and actual 4C-seq viewpoints are highlighted. (ref1)
Quick Biology is Expert and interested in genomics and high throughput screening. Find More at Quick Biology.
Ref:
1. Wu, S. et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575, 699–703 (2019).



