2020-04-12 by Quick Biology Inc.
T cell receptor (TCR) diversity or heterogeneity is a great measure of the immune response to pathogens, cancer, and therapeutic interventions. Identification of the exact pair of TCR-alpha and TCR-beta chain interacting with an antigen provides a potential vaccination therapy for cancer. Multiplex PCR amplicon sequencing is a straightforward strategy for analyzing TCR alpha and beta variables. However, non-specific amplification, primer dimers caused by multiple primers in PCR generate lots of non-specific NGS reads. Today, we introduce RNase H-dependent PCR (rhPCR) by increasing the precision of target sequence amplification while limiting primer dimers. “DNA primers that contain a single ribonucleotide residue and a 3′ blocking moiety. The blocked primers are activated when cleaved by the RNase H2 enzyme. Cleavage occurs on the 5′ side of the RNA base after primer hybridization to the target DNA. Because the primers can only be cleaved after they hybridize to the perfectly matched target sequence, primer-dimers are reduced.”(ref1, Fig 1, 2). Danna-Farber Institute used this strategy, together with adding a unique molecular identifier (UMI) to each cDNA molecule, improved the accuracy of TCR repertoire-frequency measurement (Fig.3), details please see 2019 Nature Protocols (ref2).
Figure 1: Schematic representation of rhPCR. IDT scientists have developed RNase H2–dependent PCR, a method for increasing PCR specificity and eliminating primer-dimers by using RNase H2 from Pyrococcus abyssi (P.a.) and DNA primers that contain a single ribonucleotide residue and a 3′ blocking moiety. The blocked primers are activated when cleaved by the RNase H2 enzyme. Cleavage occurs on the 5′ side of the RNA base after primer hybridization to the target DNA. Because the primers can only be cleaved after they hybridize to the perfectly matched target sequence, primer-dimers are reduced. The requirement for high target complementarity reduces the amplification of closely related sequences. (ref1)
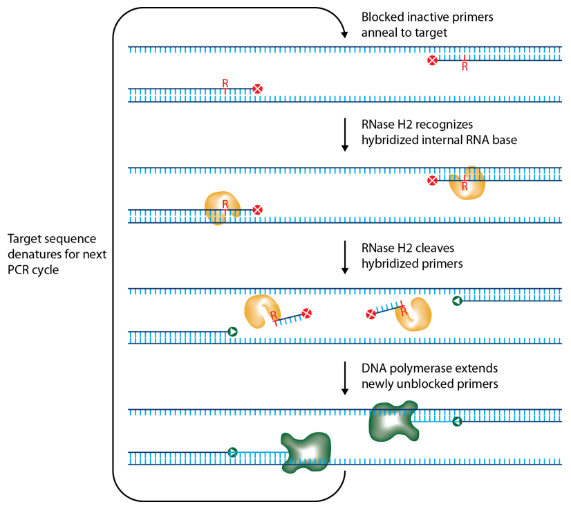
Figure 2: rhPCR primers virtually eliminate primer-dimers and nonspecific amplification artifacts in multiplex PCR (ref1)
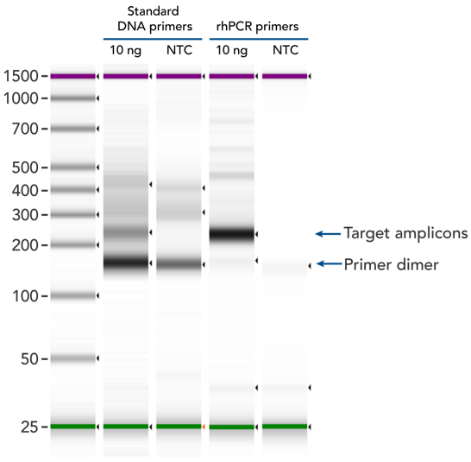
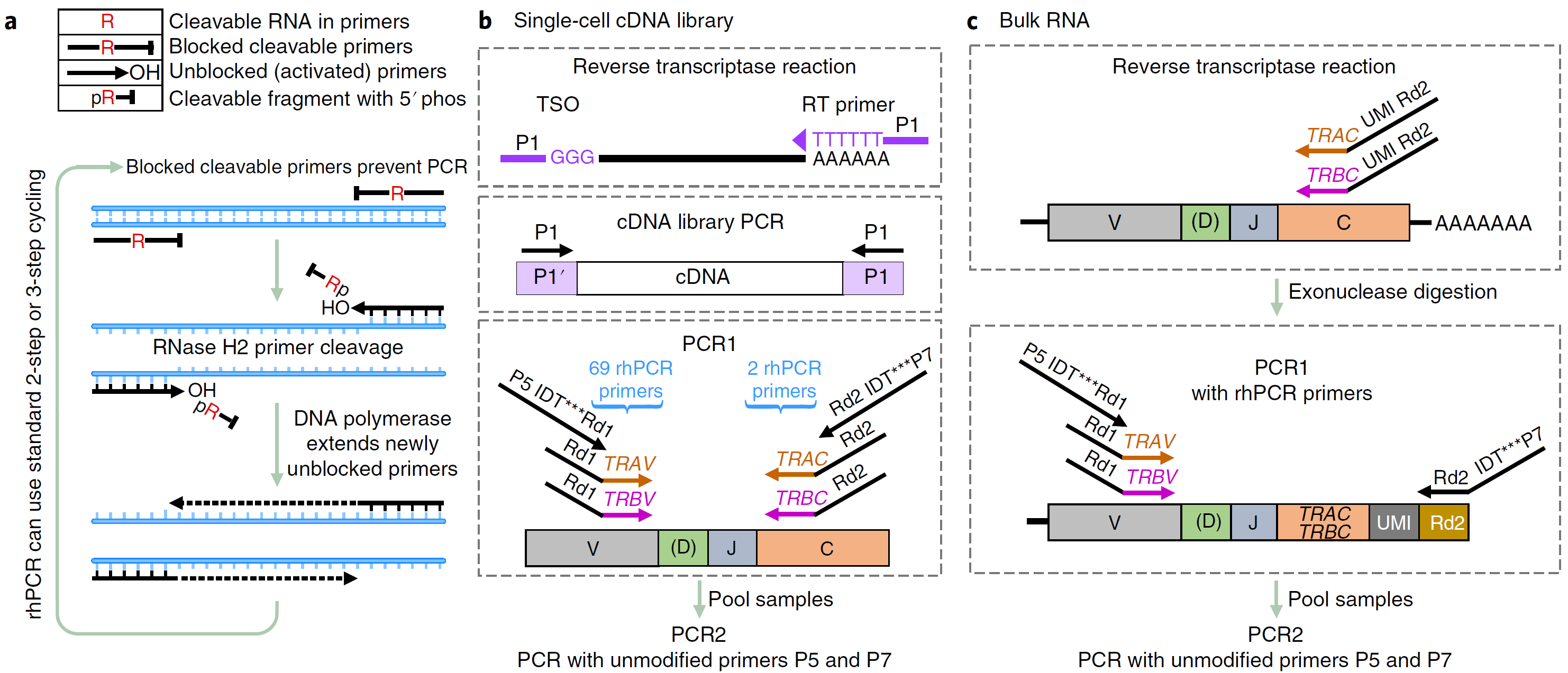
Figure 3: Features and schemes of rhTCRseq. a, Mechanism for the enhanced specificity of rhPCR. Instead of conventional primers, rhPCR uses 3ʹ-blocked oligonucleotides each containing a single ribo base. Upon high fidelity hybridization to its target, each oligonucleotide is cleaved at the ribo base by thermostable RNase H2 to generate a primer with a 3ʹ-hydroxyl that can be extended by DNA polymerase. p, phosphate group; R, ribo base. b,c, Scheme for TCR-specific amplification from single-cell cDNA libraries (b) and from bulk RNA (c). V, (D), J, and C indicate the Variable, Diversity (beta only), Joining and Constant segments of the TCR transcript, respectively. Arrowheads indicate the 3ʹ end of primers. TSO refers to the NEBNext Template Switching Oligo. P1 refers to the NEBNext Single Cell cDNA PCR Primer. Segments specific for the variable (TRAV, TRBV) and constant (TRAC, TRBC) regions of alpha and beta TCR transcripts, and for the Illumina read 1 and read 2 sequences (Rd1, Rd2) are labeled. There are 69 rhPCR primers specific for TRAV and TRBV, 1 rhPCR primer specific for TRAC, and 1 rhPCR primer specific for TRBC. IDT*** indicates 8-nt index sequence. P5 and P7 refer to the Illumina sequences required for flow cell attachment. P5 IDT***Rd1 and P7 IDT***Rd2 are rhPCR primers and refer to P5.IDTxxx. Rd1x.x1 and P7.IDTyyy.Rd2x.x1 used in the text, respectively. (Ref 2)
Quick Biology is an expert in NGS assay development. Find More at Quick Biology.
Ref:
- 1. IDT RNaseH2: https://www.idtdna.com/pages/products/qpcr-and-pcr/master-mixes-reagents/rnase-h-enzyme
- 2. Li, S. et al. RNase H–dependent PCR-enabled T-cell receptor sequencing for highly specific and efficient targeted sequencing of T-cell receptor mRNA for single-cell and repertoire analysis. Nat. Protoc. 14, 2571–2594 (2019).



