2020-09-07 by Quick Biology Inc.
Single-cell RNA sequencing is a great method to study cell heterogeneity, track cell lineages in developments, but it obscures spatial (location) and temporal resolution. To get spatial information, many other methods are developed such as (1)Geo-seq (Geographical position sequencing) through tissue staining and laser capture microdissection developed by Dr. Han and Dr.Jing’s group in CHINA (Ref1) ; (2) Slide-seq, whereby RNA was spatially resolved from tissue sections by transfer onto a surface covered with DNA-barcoded beads developed by Dr. Chen and Dr. Macosko in Broad Institute in the USA (Ref2).
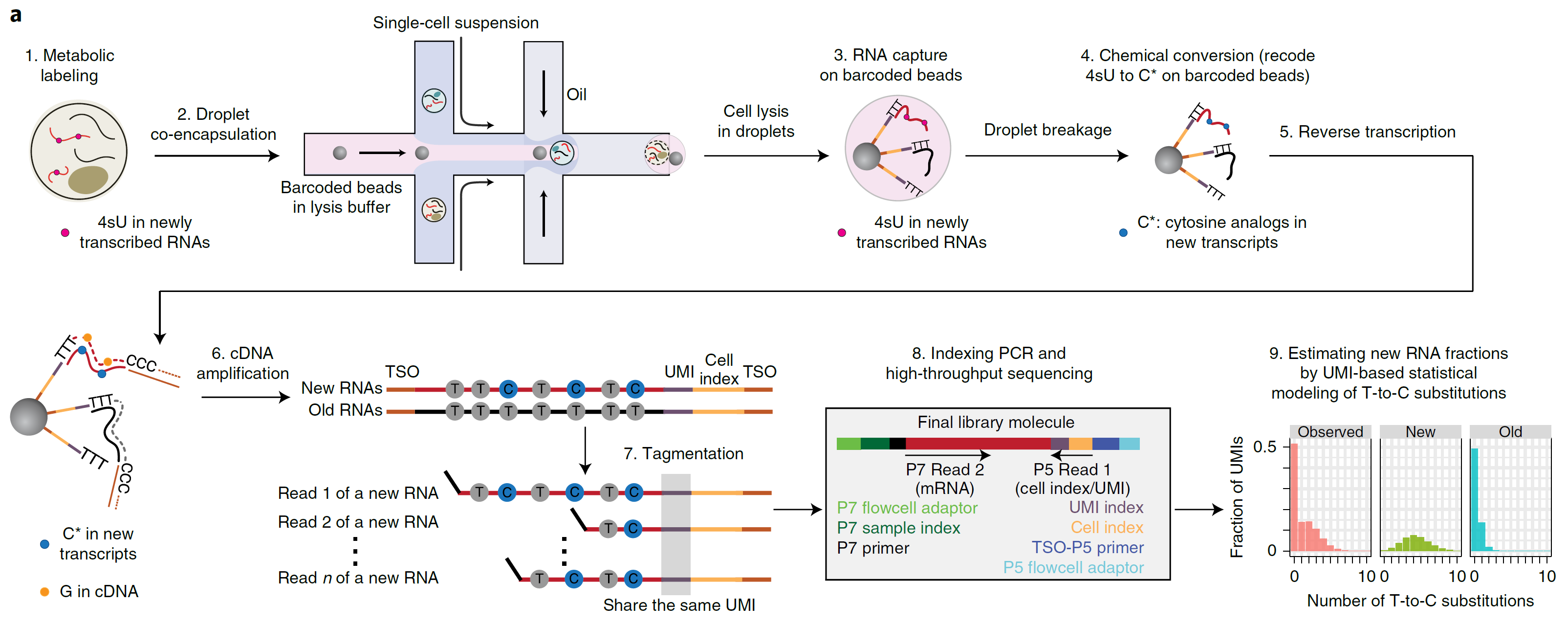
To get temporal information, in Nature Methods, Dr. Wu’s Lab developed single-cell metabolically labeled new RNA tagging sequencing called scNT-seq. Using scNT-seq, researchers can jointly profile new and old transcriptomes in single cell level (Ref3, Fig.1). Basically, the steps are:
Step1: cells are metabolically labeled with 4sU
Step2: single cell is encapsulated with barcoded oligod(T)-coated beads
Step3: mRNAs are capture by oligod(T), 4sU is chemically converted to C on beads.
Step4: reverse transcription, cDNA amplification, tagmentation and indexing PCR;
Stpe5: High-throughput sequencing and data analysis.
By scNT-seq, Dr. Wu revealed time-resolved transcription factor activities and cell-state trajectories at the single-cell level in response to neuronal activation, They further determined rates of RNA biogenesis and decay to uncover RNA regulatory strategies during stepwise conversion between pluripotent and rare totipotent two-cell embryo (2C)-like stem cell states.
Fig. 1: Overview of scNT-SEQ. TSO, template switch oligo. (image from Ref3)
Quick Biology is an expert in single-cell profiling. Find More at Quick Biology.
Ref:
- 1. Chen, J. et al. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 12, 566–580 (2017).
- 2. https://science.sciencemag.org/content/363/6434/1463
- 3. Qiu, Q. et al. Massively parallel and time-resolved RNA sequencing in single cells with scNT-seq. Nat. Methods (2020). doi:10.1038/s41592-020-0935-4



