02-15-2021 By Quick Biology
The Ubiquitin/Proteasome System (UPS) is a highly regulated mechanism of intracellular protein degradation and turnover, plays major roles in a variety of basic pathways during cell life and death. The UPS is also an attractive target for novel cancer therapies, however, the precise UPS genes and substrates important for cancer growth are not completely understood.
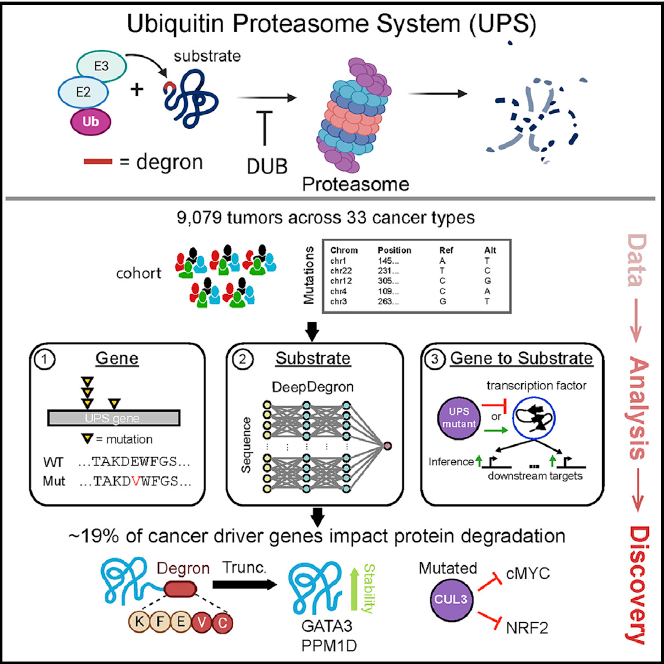
In Molecular Cell, Collin Tokheim, Xiaoqing Wang, and their colleagues comprehensively analyze multi-omics data across more than 9,000 human tumors and 33 cancer types in the TCGA cohort dataset (Fig1, Ref1). Using Tokheim et.al 2016 STAR methods, they find ~19% of cancer driver genes affect protein degradation, identify 63 unique UPS genes as putative cancer drivers. Furthermore, they identify UPS substrate mutations under positive selection in tumors by finding enriched missense mutations at known degron-related sites (i.e., the short linear amino acid motifs recognized by E3 ubiquitin ligases). Moreover, they develop a deep learning model (deepDegron) to identify mutations that result in degron loss. Excitingly, the researchers use their model, predict that gain-of-function truncating mutations in GATA3 and PPM1D result in increased protein stability. This study demonstrates the important role of UPS dysregulation in human cancer and underscores the potential therapeutic utility of targeting the UPS.
Figure 1: Graphical Abstract of UPS mutations in human cancers.
Quick Biology can assist you with RNA-seq and TCGA data analysis. Find More at Quick Biology.
See resource:
1. Tokheim, C. et al. Article Systematic characterization of mutations altering protein degradation in human cancers Article Systematic characterization of mutations altering protein degradation in human cancers. Mol. Cell 1–17 (2021). doi:10.1016/j.molcel.2021.01.020



