Introduction
Quickly Targeting and Validating 100 Pathways for Gene-Drug and Drug-Drug Interaction Studies
The 100-QuickPathTM Drug Array is a technological innovation for conducting functional studies on your pathway of interest through drug-drug and drug-gene interactions. Validated drugs for 22 key pathway-focused genes with appropriate controls are arrayed on a 96-well plate. The drugs in the plate are highly selective to their targets. Following a simple cell culture protocol with the drug in the plate, you can quickly and directly determine the phenotype, cell death and cytotoxicity of cells on the same plate with colorimetric, luminescent, or fluorescent cell-based detection methods. The ultimate goal of the array is to quickly enable you to screen and validate the interactions of the candidate gene/drug with 100 targets/pathways in 3 days.
Order Information:
Please check, download the Drug Array manual and analysis template here:
![]() Drug Array data analysis template v4.xlsx
Drug Array data analysis template v4.xlsx
Please click the corresponding Drug array product to see the detailed plate design and gene targets.
Why Use Drug Arrays?
100-QuickPathTM Drug Array Applications
1. Drug-Drug interaction
The strategy is to find best drug combinations to overcome drug resistance by blocking the original and rewired pathways. The 100-QuickPathTM Drug Array screening also allows identifying new mechanisms for a drug candidate.
Example 1: MEK inhibitor (GSK1120212) & CDK4 Inhibitors (PD-0332991) in SB-2 cell line.
Melanomas with a mutant NRAS oncogene have no effective targeted therapy. Research indicated that Cdk4 protein as the key effector for tumor progression. PD-0332991, a selective dual inhibitor of Cdk4 and 6, was then used in combination with the MEK inhibitor GSK1120212 to mimic the effects of NRASQ61K extinction and tumor regression was achieved in SB-2 tumor xenograft [1].
Example 2: RAF Inhibitors (PLX4032) and EGFR inhibitor (BIBW2992) in HTC-C3 cell line
The BRAF oncogene has activating mutations in some cancers, and the inhibitor PLX4032 exhibits a high success rate in melanomas but not in colorectal cancers. A kinase screening assay using a library of 535 shRNAs revealed a potential for the inhibition of EGFR to synergize with BRAF inhibition. Further investigation showed that the inhibition of BRAF causes strong feedback activation of EGFR, leading to poor therapeutic efficacy in colorectal cancers with BRAF mutations. This provides a strong rationale for the combined use of BRAF and EGFR inhibitors. Moreover, the combined use of BRAF and EGFR inhibitors induced apoptosis, but either drug alone could not produce the same effect. This discovery was made possible through a large kinase screening and validation project which required significant costs and efforts [2].
2. Gene-Drug interaction
Expression of certain genes may render cancer cell sensitive or resistance to anti-cancer drugs. The 100-QuickPathTM Drug Array can evaluate how your candidate gene interacts with known drugs, and reveal synthetic lethality or biomarkers for drug sensitivity.
Example 1: PARP inhibitor (AZD2281) and EWS-FLI1 siRNA in A673 cell line
Targeted cancer therapy, such as the inhibitor of the BCR-ABL translocation gene product, has been a major breakthrough in the field and the mechanism of action has been well studied. Although there are a number of cancer drugs in the clinic and under investigation, the responsiveness is widely variable and the mechanisms are poorly understood. There is also a lack of biomarkers to predict their therapeutic effectiveness. To find new biomarkers for these cancer drugs, a large-scale screening study was carried out and 48,178 drug–cell-line combinations were assayed. Most of the cancer genes and drugs had at least one interaction associated with sensitivity or resistance. The EWS-FLI1 rearrangement is an example of one such biomarker for sensitivity to PARP inhibitors in Ewing’s sarcoma cells [3].
In this drug assay set, we have 66 chemicals in 3 arrays, selectively targeting 66 primary targets. The targets of these chemicals are well defined and the chemicals are well characterized. Therefore, through our Drug Array platform, the effects of the drug combination and mechanisms of gene-drug interactions can be interrogated at a much lower cost and effort. Quick Biology 100-QuickPathTM Drug Arrays aims to screen the interaction of the candidate gene/drug with as many as 100 targets/pathways in 3 days.
How does it work?
--Simple Experimental Designs and Workflow:
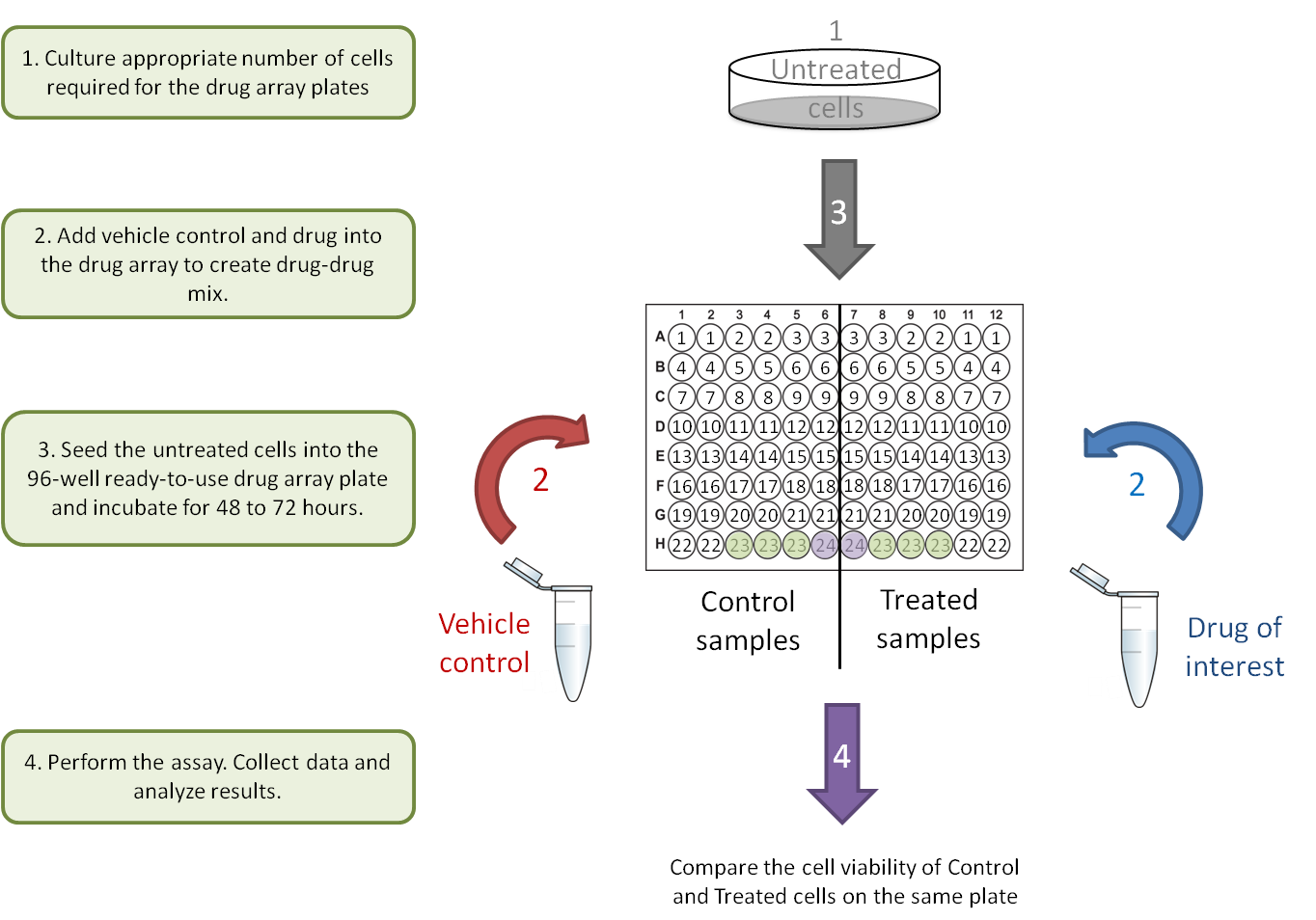
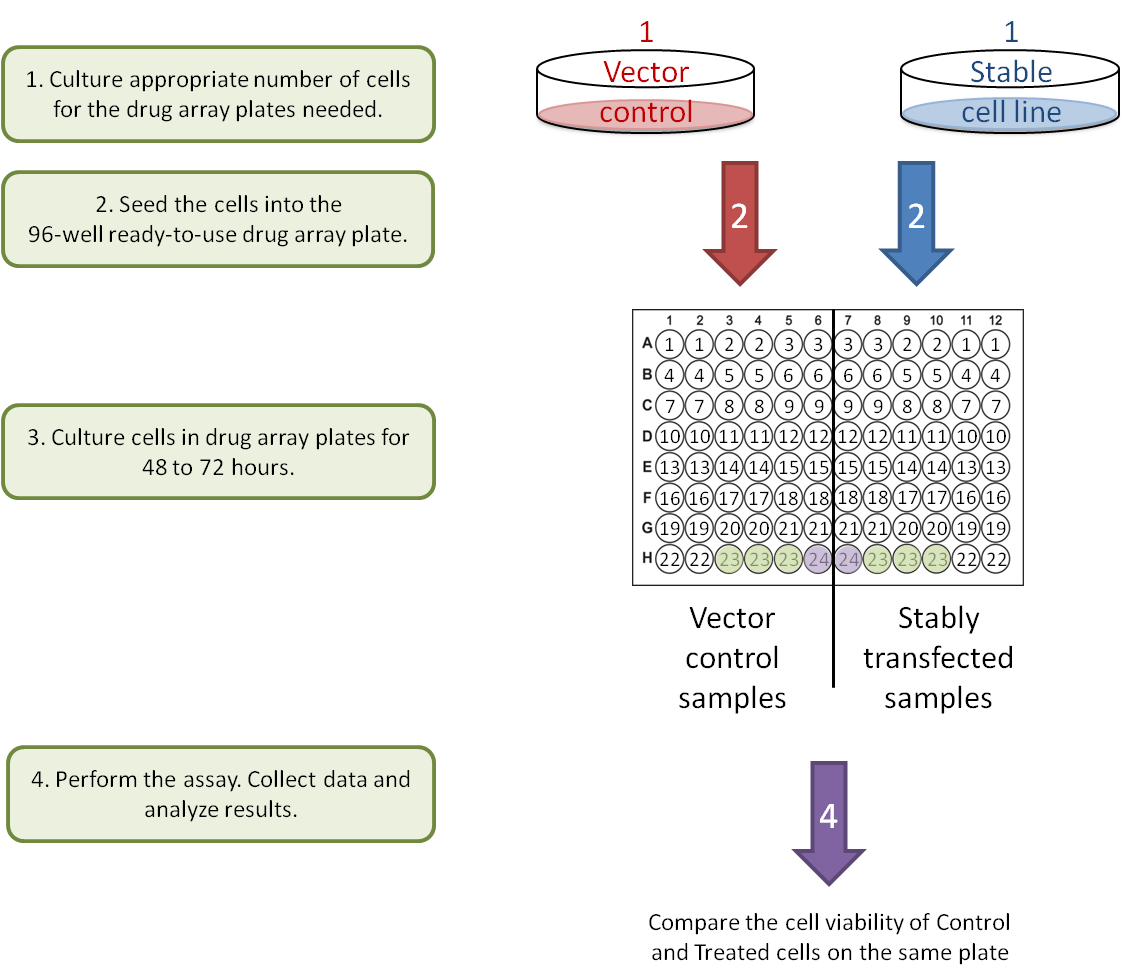
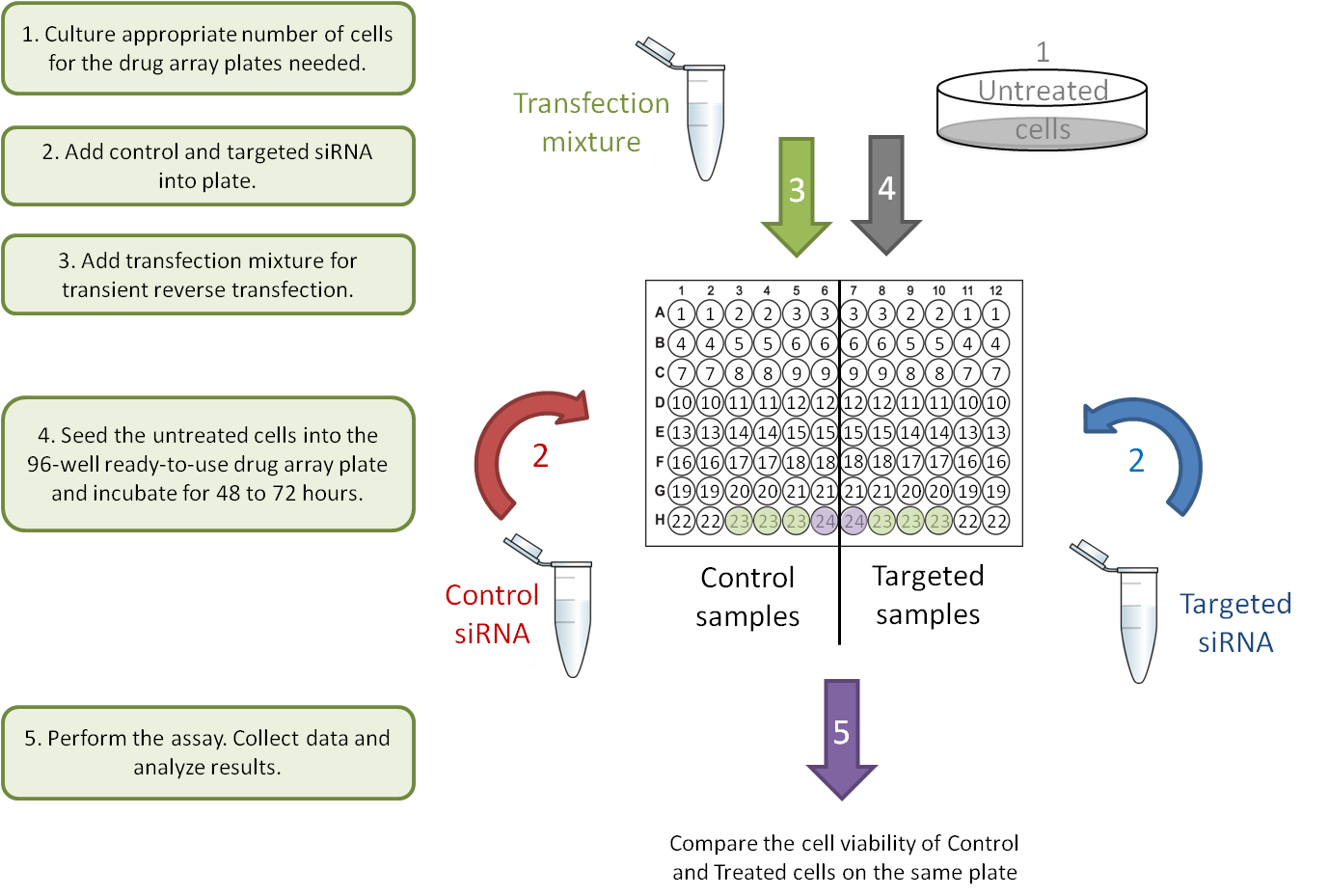
Each Drug Array plate is designed to compare two different samples in parallel on one ready-to-use 96-well plate. To better observe the drug-drug and gene-drug interactions, each drug is provided at two concentrations (0.5 µM and 2.5 µM). This concentration range has been optimized to block the activity of the drug target in most cell lines. Following a simple cell culture protocol with the drug in the plate, you can quickly and directly determine the phenotype, cell death and cytotoxicity of cells on the same plate with colorimetric, luminescent, or fluorescent cell-based detection methods.
Please find Drug Array product manual here.
A. Drug-gene interaction.
B. Gene-Gene interaction: stable cell lines.
C. Gene-Gene interaction: transient transfections.
-- Ready-to-use data analysis:
Quick Biology presents a user-friendly analysis template in excel format. There are separate analysis templates for Drug Array I, II, and III, respectively.
![]() Drug Array data analysis template v4.xlsx
Drug Array data analysis template v4.xlsx
Users will only need to input their own data into the Raw Data sheet. The pre-programmed algorithm on the Analysis sheet will subtract the background (plate well # 24), normalize against the DMSO control (plate well # 23), and calculate the average, standard deviation, inhibition rate and ratio of inhibition for each treatment and control pair. The results will be transformed into figures for easy interpretation.
Reference
(1) Kwong LN, Costello JC, Liu H, Jiang S, Helms TL, Langsdorf AE, Jakubosky D, Genovese G, Muller FL, Jeong JH et al: Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nature medicine 2012, 18(10):1503-1510.
(2) Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R: Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR.Nature 2012, 483(7387):100-103.
(3) Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J et al: Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483(7391):570-575.



